-
PDF
- Split View
-
Views
-
Cite
Cite
Ting Chen, Zhouqing Huang, Liansheng Wang, Yue Wang, Feizhen Wu, Shu Meng, Changqian Wang, MicroRNA-125a-5p partly regulates the inflammatory response, lipid uptake, and ORP9 expression in oxLDL-stimulated monocyte/macrophages, Cardiovascular Research, Volume 83, Issue 1, 1 July 2009, Pages 131–139, https://doi.org/10.1093/cvr/cvp121
Close - Share Icon Share
Abstract
The inflammatory responses of monocytes/macrophages and the stimulation of lipid uptake into these cells by oxidized low density lipoprotein (oxLDL) are critical to the initiation and development of atherosclerosis. Increasing evidence has demonstrated that many microRNAs play important roles in the cell proliferation, apoptosis, and differentiation that accompany inflammatory responses. However, whether microRNAs are associated with monocyte/macrophage inflammatory responses or oxLDL stimulation is not yet known. The aim of the present study is to investigate microRNAs in monocytes/macrophages and their potential role in oxLDL-stimulation of lipid uptake and other atherosclerotic responses.
Microarrays were used to analyse the global expression of microRNAs in oxLDL-stimulated human primary peripheral blood monocytes. Expression profiles of the microRNAs were verified using TaqMan real-time PCR. Five microRNAs (microRNA-125a-5p, microRNA-9, microRNA-146a, microRNA-146b-5p, and microRNA-155) were aberrantly expressed after oxLDL treatment of human primary monocytes. Bioinformatics analysis suggested that microRNA-125a-5p is related to a protein similar to ORP9 (oxysterol binding protein-like 9) and this was confirmed by a luciferase reporter assay. MicroRNA-125a-5p was found to mediate lipid uptake and to decrease the secretion of some inflammatory cytokines (interleukin-2, interleukin-6, tumour necrosis factor-α, transforming growth factor-beta) in oxLDL-stimulated monocyte-derived macrophages.
MicroRNA-125a-5p may partly provide post-transcriptional regulation of the proinflammatory response, lipid uptake, and expression of ORP9 in oxLDL-stimulated monocyte/macrophages.
1. Introduction
Atherosclerosis is a chronic inflammatory disease of the arterial wall with enormous epidemiological relevance1,2 and monocytes/macrophages play important roles in the formation of atherosclerotic lesions.3,4 Circulating monocytes migrate under the subendothelial space and differentiate into macrophages as one of the key steps in the development of atherosclerosis. Macrophages then take up oxidized low density lipoprotein (oxLDL), which leads to their conversion into foam cells,5–7 and these foam cells can in turn secrete many proinflammatory factors, such as transforming growth factor (TNF)-α and interleukins (ILs).8 Cholesterol loaded macrophage foam cells are the hallmark of early atherosclerosis and eventually undergo secondary necrosis to form the lipid core of advanced atherosclerotic plaques. When exposed by rupture or erosion, the core triggers acute thrombotic events leading to myocardial infarction and strokes.9
In addition, oxLDL has long been known to contain lipids with chemotactic activity toward human monocytes, therefore, its accumulation also increases monocyte recruitment to the vascular wall.10 Thus, monocyte/macrophage involvement in atherosclerosis appears to involve a range of functions, including the expression of multiple proinflammatory factors, adhesion molecules, chemotactic factors, and scavenger receptors that are regulated by oxLDL-stimulation.11 However, little is known regarding the complex upstream regulators of gene expression and translation involved in these responses.
MiRNAs, an emerging class of highly conserved, non-coding small RNAs, regulate gene expression at the post-transcriptional level by inhibiting the translation of protein from mRNA or by promoting the degradation of mRNA. MiRNAs are transcribed by RNA polymerase II as part of a primary transcript12,13 that is processed by Drosha and DGCR8 into a smaller RNA molecule.14 Mature miRNAs specifically bind to 3′-UTRs of target cellular mRNAs, leading to either mRNA degradation or inhibition of translation.15 More than 500 human miRNAs have been identified so far, and increasing evidence indicates that miRNAs have distinct expression profiles and play essential roles in various physiological and pathological processes, including cardiogenesis, haematopoietic lineage differentiation, and oncogenesis.16
A few specific miRNAs that regulate endothelial cell functions and angiogenesis have been described.16 For example, Let7-f, miR-27b, and miR-130a have been identified as pro-angiogenic miRNAs. Others, such as miR-221 and miR-222, have been shown to inhibit in vitro endothelial cell migration, proliferation, and angiogenesis.16 Early studies also indicate that specific miRNAs (e.g. miR-155, miR-21, and miR-126) are responsible for vascular inflammation and diseases.16 Thus, the identification of miRNAs and their respective targets may offer new therapeutic strategies for the treatment of a number of vascular diseases such as atherosclerosis, for improvement of neovascularization after ischaemia, or for the prevention of atherosclerotic inflammation. Therefore, the aim of the present study was to use a combination of microarray and TaqMan real-time PCR to analyse miRNA expression profiles in oxLDL-stimulated human primary peripheral blood monocytes, to study the possible roles of miRNAs in atherosclerotic processes in these cells.
2. Methods
The investigation conformed with the principles outlined in the Declaration of Helsinki for use of human blood and was approved by the Ethics Committee of Experimental Research, JiaoTong University Shanghai Medical College.
2.1 Human primary peripheral blood monocyte isolation and culture with oxLDL
Peripheral human blood was obtained from healthy donors. Mononuclear cells were isolated by centrifugation through a Ficoll-isopaque (Sigma, St Louis, MO, USA) density gradient.17 To obtain the monocytes, the mononuclear cells were allowed to adhere to six-well plates with 5% autologous serum for 2 h at 37°C, in a 5% CO2 incubator. Non-adherent cells were removed and the adherent cells were co-cultured with oxLDL (30 μg mL−1) for 6–12 h.
2.2 Analysis of the expression of miRNAs by μParaflo™ MicroRNA microarray assay
Total RNA was extracted from monocytes using Trizol reagent according to the manufacturer's instructions (Qiagen kit). Microarray assay was performed with the μParaflo™ MicroRNA microarray assay system (LC Sciences). The assay was initiated with 2–5 µg total RNA, and the small RNAs (<300 nt) isolated were 3′-extended with a poly(A) tail using poly(A) polymerase. Thereafter, an oligonucleotide tag was ligated to the poly(A) tail for later fluorescent dye staining; two different tags were used for the different RNA samples. The detection probes were made by in situ synthesis using photogenerated reagent chemistry. The detection used fluorescence labelling by tag-specific Cy3 and Cy5 dyes, whereas the images were collected using a laser scanner (GenePix 4000B, Molecular Device) and were digitized using the Array-Pro image analysis software (MediaCybernetics). Each miRNA probe was repeated in septa-replicate on the chip. Microarray data were analysed by first subtracting the background (the median of 5–25% of the lower signal intensities) and then the signal was normalized using a LOWESS (locally weighted regression) filter.18 For each sample, triplicates were analysed at each time point and significant differences between 0, 6, 12 h for a given detectable miRNA signal were calculated. The ratio of the two sets of detected signals (log 2 transformed, balanced) and the P-values of the t-test and ANOVA were calculated. The differentially detected signals with P < 0.05 were analysed using gene hierarchical clustering of the log 2 value of each different time group and then were displayed in a heatmap. Clustering was performed using Cluster 3.0 created by Michiel de Hoon, Seiya Imoto, and Satoru Miyano, University of Tokyo, Human Genome Center (Euclidean distance, links using the average) and viewed in heatmap using Java TreeView 1.0.13 software.
2.3 miRNA real-time quantitative PCR
As determined by the microarray results, the five most notable aberrantly expressed microRNAs (miR-125a-5p, miR-9, miR-146a, miR-146b-5p, and miR-155) were further measured using TaqMan real-time PCR. The PCR reaction was directly monitored by the ABI PRISM 7000 Sequence Detection System (Applied Biosystems, CA, USA). Briefly, cDNA was made from enriched miRNA using the TaqMan MicroRNA RT kit. U6 RNA was used as an endogenous control.
2.4 THP-1 cell culture and anti-miRNA transfection
The human monocytic cell line THP-1 was obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA). This was maintained at a density of 106/mL in RPMI 1640 medium supplemented with 10% foetal bovine serum, 10 mM HEPES (Sigma), 1% pen/strep solution, and incubated in a 5% CO2 incubator at 37°C. To induce monocyte differentiation to macrophages, THP-1 cells were cultured with 100 nM phorbol 12-myristate 13-acetate (PMA) (Calbiochem, San Diego, CA, USA) for 24 h.19
MiR-125a-5p inhibitor (Ambion) was transfected into PMA-induced THP-1 using the fast transfection protocol recommended by Qiagen. The miR-125a-5p inhibitor was diluted in the medium at a final concentration of 50 nM. To confirm the efficiency of transfection, the same amount of Cy3-labelled negative control (Ambion) was also transfected. Cells were incubated with miR-125a-5p inhibitor for 24 h and then exposed to oxLDL (50 μg mL−1) for 24 h.
2.5 HPLC analysis of the lipid levels
PMA-differentiated THP-1 cells were transfected with miRNA inhibitor, oxLDL was added 24 h after transfection, and cells were further incubated for 24 h. The sterol analyses were performed using a high performance liquid chromatography (HPLC) system (model 2790, controlled with Empower Pro software; Waters Corp., Milford, MA, USA). Sterols were detected using a photodiode array detector equipped with a 4 µL cell (model 996; Waters Corp.). Analysis of cholesterol and cholesteryl esters was performed after elution with acetonitrile-isopropanol 30:70 (v/v)20 and detection by absorbance at 210 nm.
2.6 Flow cytometry
PMA-differentiated THP-1 cells were transfected with miRNA inhibitor for 24 h, then fluorescent-tagged Dil-oxLDL was added, and the cells were incubated for 24 h. Adherent cells were harvested, washed once with phosphate buffer saline, and counterstained immunophenotypically for anti-CD68, anti-CD36, anti-LOX-1. Analysis was performed on a fluorescent activated cell sorting (FACS) calibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA) with Cell Quest Pro software.
2.7 ELISA assays of inflammatory markers
PMA-differentiated THP-1 cells were transfected with miRNA inhibitor for 24 h, then cells were further incubated with oxLDL for another 24 h. Culture supernatants were analysed to determine TNF-α, IL-6, IL-2, and TGF-β using Sandwich Enzyme Immunoassay kits (R&D Systems Europe Ltd, Abingdon, UK) according to the manufacturer's instructions.
2.8 Cloning of 3′-UTR of ORP9 mRNA and reporter gene assay
Expression sequence tag clones containing the 3′-UTR sequences from ORP9 cDNA were obtained commercially (Invitrogen) and used as templates for PCR; ORP9 3′-UTR primers spanned +2231/2919 (GenBank accession no. NM_024586). The amplified PCR product was gel purified and subcloned into pGL3 (Promega). The insert was excised with Fast Digest HindIII and SacI restriction enzymes (Fermentas, Hanover, MD, USA), gel purified, and ligated into the pGL3 miRNA luciferase reporter vector (Promega). At 50% confluence, THP-1 was cotransfected with 100 ng p-ORP9 UTR miRNA luciferase reporter vector and anti-miR-125a-5p (50 nM) using the Hiperfect transfection reagent (Qiagen). Cells were also transformed with 100 ng PGL3-control vector, which is useful for monitoring transfection efficiency. Use anti-miR negative, a non-targeting negative control. All cells were also transfected with pRL-TK (Promega) for normalization control. After 48 h, cells were washed and lysed with Passive Lysis Buffer, and firefly luciferase activities were determined using the dual-luciferase reporter assay system and a luminometer. The relative reporter activity was obtained by normalization to the Renilla control luciferase activity.
2.9 mRNA real-time quantitative PCR and western blot analysis
PMA-differentiated THP-1 cells were transfected with miRNA inhibitor, oxLDL was added 24 h after transfection, and cells were further incubated for 24 h. mRNA levels were analysed using the SYBR Green reagent kits with gene specific primers on the Applied Biosystems 7000 real-time PCR system, according to the manufacturer's instructions. An ORP9-specific 483 bp fragment was amplified using a forward primer (GAAATAGGCGAGTGACGTCAG; nucleotides −50 to −30) and a reverse primer (GAAGCTTGTCATCAAAAAGCTTTAATTGTTC; nucleotides 403–435). As an internal standard, human glyceraldehyde 3-phosphate dehydrogenase was also amplified. The protein extracts were denatured and the solubilized proteins (20 μg) subjected to electrophoresis on 10% polyacryl amide SDS gels, then transferred onto polyvinylidene difluoride membranes (Millipore, MA, USA). This was followed by probing with primary antibodies for rabbit anti-human-ORP9 (diluted 1:1000 in TBST), or rabbit anti-actin (diluted 1:5000 in TBST) for 2 h, and then by goat anti-rabbit secondary antibody labelled with far-red-fluorescent Alexa Fluor 680 dye. All signals were detected by Odyssey (Li-cor, USA). Densitometric analysis was performed by using the Quantity One (Bio-Rad) to scan the signals.
2.10 Statistical analysis
Data are expressed as mean ± SD. Differences were compared by one-way ANOVA. A value of P < 0.05 was considered statistically significant. All experiments were performed at least three times.
3. Results
3.1 Up/down-regulated miRNAs in oxLDL-stimulated monocytes
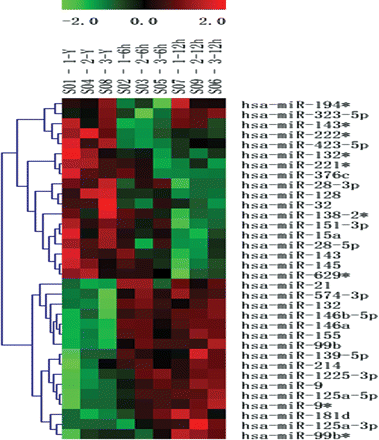
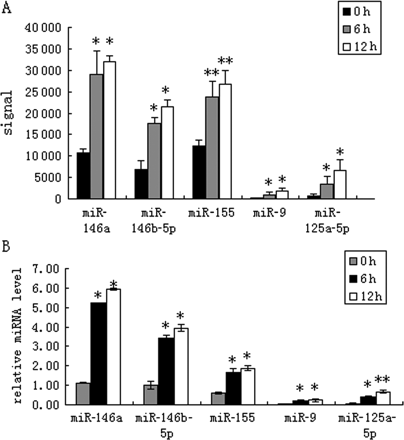
In order to identify the up/down-regulated miRNAs in oxLDL-stimulated monocytes, we analysed the expression profile of miRNAs in oxLDL-stimulated human primary monocytes using microarrays that contained 723 oligonucleotide probes complementary to mature forms of miRNAs of human, mouse, and rat origin, based on version 10.1 of the Sanger miRBase (http://microrna.sanger.ac.uk/sequences). Microarray analysis showed an aberrant miRNA expression in oxLDL-stimulated cells. After normalizing the microarray values using LOWESS filter, values with significant differences (P < 0.05) were analysed using hierarchical clustering of the log 2 value and displayed in a heatmap (Figure 1). These results reflect the temporal changes of miRNA expression levels during the monocyte response to oxLDL stimulation. OxLDL significantly up-regulated several miRNAs (e.g. miR-146a, miR-155) as depicted by the shades of red in the heatmap (P < 0.05, Figure 1). OxLDL also down-regulated some miRNAs (e.g. miR-128, miR-15a), which are shown as shades of green in the heatmap. The five miRNAs (miR-125a-5p, 9, 146a, 146b-5p, 155) with the highest fold changes are listed in Figure 2A. The raw data for the microarray are shown in Supplementary material online. To verify the accuracy of the microarray results, we selected five miRNAs (miR-125a-5p, 9, 146a, 146b-5p, 155) for further identification using a TaqMan real-time quantitative PCR. Similar results were obtained by TaqMan real-time quantitative PCR of the five miRNAs (Figure 2B) and indicated that miR-125a-5p expression had increased more than 11-fold. Thus, miR-125a-5p was further studied to determine its potential biological function.
Dysregulation of miRNA expression in monocytes after oxLDL stimulation. Differentially expressed miRNAs (P < 0.05) were analysed by hierarchical clustering of the log 2 value of each miRNA microarray signal at 0, 6, and 12 h. Red: up-regulation; green: down-regulation; black: no change. The legend on the right displays the microRNA represented in the corresponding row. The bar code on the top represents the colour scale of the log 2 values. Each column represents the data from a given time course and includes three repetitions at the top of the heatmap.
Differentially expressed miRNAs in oxLDL treated-human primary monocytes. The five most notable aberrantly expressed miRNAs were also identified by TaqMan real-time PCR. (A) The signal value of the five miRNAs with highest-fold change. *P < 0.05, **P < 0.01 compared with 0 h group. (B) Total RNA was isolated from freshly isolated monocytes. Relative expression levels of miRNA-125a-5p, miRNA-9, miRNA-146a, miRNA-146b-5p, and miRNA-155 were analysed using the TaqMan miRNA assay system. Data are expressed as mean ± SD (n = 3), *P < 0.01 compared with 0 h group, **P < 0.01 compared with *marked group.
3.2 Effects of miR-125a-5p inhibitor on lipid uptake and expression of surface scavenger receptors in oxLDL-stimulated macrophages
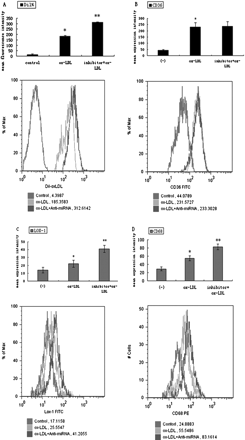
After the treatment with oxLDL and miR-125a-5p inhibitor, the levels of cholesterol in PMA-differentiated THP-1 cells were assessed by HPLC. As shown in Table 1, the level of total and esterified cholesterol increased significantly in the group with oxLDL and miR-125a-5p inhibitor (25 or 50 nM) compared with the cells treated with oxLDL only. Moreover, when we substituted Dil-labelled-oxLDL for oxLDL, and analysed the mean fluorescence intensity of Dil-oxLDL in oxLDL-stimulated macrophages by FACS, similar results were obtained, as shown in Figure 3A. These result suggested that miR-125a-5p could decrease the lipid uptake in oxLDL-stimulated monocyte/macrophages.
Effects of miR-125a-5p inhibitor on macrophage phagocytosis and surface scavenger receptors. THP-1 cells were incubated in the absence or presence of the miR-125a-5p inhibitor or oxLDL for 48 h. (A) Mean fluorescence intensity of Dil-LDL (%), (B) expression of CD36, (C) expression of LOX-1, (D) expression of CD68. Values (Mean ± SD) are calculated from five different experiments, *P < 0.05 vs. control; **P < 0.05 vs. *marked groups.
Effects of miR-125a-5p inhibitor on the level of total, free, and esterified cholesterol in THP-1 macrophage-derived foam cells
| . | Total cholesterol . | Free cholesterol . | Esterified cholesterol . | CE/TC% . |
|---|---|---|---|---|
| ox | 446.3 ± 11.5 | 185.7 ± 11.1 | 260.6 ± 1.5 | 58.4 ± 1.23% |
| ox + 25 nM | 573 ± 11* | 192.7 ± 7.5* | 380.3 ± 3.5* | 66.4 ± 0.66% |
| ox + 50 nM | 594 ± 10.5* | 186.4 ± 10.0* | 407.6 ± 1.5* | 68.6 ± 1.13% |
| . | Total cholesterol . | Free cholesterol . | Esterified cholesterol . | CE/TC% . |
|---|---|---|---|---|
| ox | 446.3 ± 11.5 | 185.7 ± 11.1 | 260.6 ± 1.5 | 58.4 ± 1.23% |
| ox + 25 nM | 573 ± 11* | 192.7 ± 7.5* | 380.3 ± 3.5* | 66.4 ± 0.66% |
| ox + 50 nM | 594 ± 10.5* | 186.4 ± 10.0* | 407.6 ± 1.5* | 68.6 ± 1.13% |
TC, total cholesterol; FC, free cholesterol; CE, esterified cholesterol. Values are represented as (mean ± SD) mg/g, n = 3.
*P < 0.01 vs. the pure ox-LDL group.
Effects of miR-125a-5p inhibitor on the level of total, free, and esterified cholesterol in THP-1 macrophage-derived foam cells
| . | Total cholesterol . | Free cholesterol . | Esterified cholesterol . | CE/TC% . |
|---|---|---|---|---|
| ox | 446.3 ± 11.5 | 185.7 ± 11.1 | 260.6 ± 1.5 | 58.4 ± 1.23% |
| ox + 25 nM | 573 ± 11* | 192.7 ± 7.5* | 380.3 ± 3.5* | 66.4 ± 0.66% |
| ox + 50 nM | 594 ± 10.5* | 186.4 ± 10.0* | 407.6 ± 1.5* | 68.6 ± 1.13% |
| . | Total cholesterol . | Free cholesterol . | Esterified cholesterol . | CE/TC% . |
|---|---|---|---|---|
| ox | 446.3 ± 11.5 | 185.7 ± 11.1 | 260.6 ± 1.5 | 58.4 ± 1.23% |
| ox + 25 nM | 573 ± 11* | 192.7 ± 7.5* | 380.3 ± 3.5* | 66.4 ± 0.66% |
| ox + 50 nM | 594 ± 10.5* | 186.4 ± 10.0* | 407.6 ± 1.5* | 68.6 ± 1.13% |
TC, total cholesterol; FC, free cholesterol; CE, esterified cholesterol. Values are represented as (mean ± SD) mg/g, n = 3.
*P < 0.01 vs. the pure ox-LDL group.
To study the potential mechanism of miR-125a-5p inhibitor effects on lipid uptake by macrophages, we analysed some associated scavenger receptors (e.g. LOX-1, CD68, CD36) in oxLDL-stimulated macrophages. MiR-125a-5p inhibitor caused enhanced expression of LOX-1 and CD68, but amounts to no more than one-third of total oxLDL taken up by macrophages, and CD36 expression showed no similar change (Figure 3B–D).
3.3 MiR-125a-5p inhibitor increased the secretion of inflammatory cytokines
The inflammatory molecules (IL-6, IL-2, TGF-β, and TNF-α) from PMA-differentiated THP-1 cell exposed to miR-125a-5p inhibitor (25, 50 nM) only, oxLDL only, and oxLDL with miR-125a-5p inhibitor were measured. As shown in Table 2, miR-125a-5p inhibitor significantly increased the secretion of inflammatory cytokines in cells that were unstimulated (only with miRNA inhibitor) or stimulated (with addition of oxLDL). The changes in TGF-β were most striking, showing a nearly three-fold increase in the anti-miR(50 nM) group compared with the control. In addition, a one- to two-fold rise in IL-6, IL-2, and TNF-α level were also observed.
Comparison of inflammatory biomarkers secreted by transfecting the miR-125a-5p inhibitor
| . | IL-6 (pg/mL) . | TNF-α (ng/mL) . | TGF-β (pg/mL) . | IL-2 (ng/mL) . |
|---|---|---|---|---|
| Mean ± SD | Mean ± SD | Mean ± SD | Mean ± SD | |
| Sham | 48.9 ± 0.80 | 1.0 ± 0.08 | 25.63 ± 0.91 | 2.8 ± 0.02 |
| anti-miR (25 nM) | 60.15 ± 0.35* | 1.075 ± 0.005* | 51.43 ± 0.64* | 4.29 ± 0.03* |
| anti-miR (50 nM) | 66.3 ± 0.52** | 1.23 ± 0.005** | 70.7 ± 0.95** | 5.09 ± 0.075** |
| Sham | 50.9 ± 0.50 | 1.15 ± 0.02 | 32.06 ± 0.46 | 3.35 ± 0.02 |
| Ox | 64.8 ± 0.35* | 1.29 ± 0.008* | 56.63 ± 0.75* | 4.46 ± 0.03* |
| anti-miR + ox | 77.65 ± 0.40** | 1.49 ± 0.017** | 75.06 ± 1.09** | 5.68 ± 0.02** |
| . | IL-6 (pg/mL) . | TNF-α (ng/mL) . | TGF-β (pg/mL) . | IL-2 (ng/mL) . |
|---|---|---|---|---|
| Mean ± SD | Mean ± SD | Mean ± SD | Mean ± SD | |
| Sham | 48.9 ± 0.80 | 1.0 ± 0.08 | 25.63 ± 0.91 | 2.8 ± 0.02 |
| anti-miR (25 nM) | 60.15 ± 0.35* | 1.075 ± 0.005* | 51.43 ± 0.64* | 4.29 ± 0.03* |
| anti-miR (50 nM) | 66.3 ± 0.52** | 1.23 ± 0.005** | 70.7 ± 0.95** | 5.09 ± 0.075** |
| Sham | 50.9 ± 0.50 | 1.15 ± 0.02 | 32.06 ± 0.46 | 3.35 ± 0.02 |
| Ox | 64.8 ± 0.35* | 1.29 ± 0.008* | 56.63 ± 0.75* | 4.46 ± 0.03* |
| anti-miR + ox | 77.65 ± 0.40** | 1.49 ± 0.017** | 75.06 ± 1.09** | 5.68 ± 0.02** |
Concentration of inflammatory cytokines secreted in 48 h culture supernatants by stimulated PMA-induced THP-1 cells from anti-miR (25 and 50 nM), ox-LDL and ox-LDL with anti-miR.
*P < 0.01 compared with control cell (sham).
**P < 0.01 compared with *group.
Comparison of inflammatory biomarkers secreted by transfecting the miR-125a-5p inhibitor
| . | IL-6 (pg/mL) . | TNF-α (ng/mL) . | TGF-β (pg/mL) . | IL-2 (ng/mL) . |
|---|---|---|---|---|
| Mean ± SD | Mean ± SD | Mean ± SD | Mean ± SD | |
| Sham | 48.9 ± 0.80 | 1.0 ± 0.08 | 25.63 ± 0.91 | 2.8 ± 0.02 |
| anti-miR (25 nM) | 60.15 ± 0.35* | 1.075 ± 0.005* | 51.43 ± 0.64* | 4.29 ± 0.03* |
| anti-miR (50 nM) | 66.3 ± 0.52** | 1.23 ± 0.005** | 70.7 ± 0.95** | 5.09 ± 0.075** |
| Sham | 50.9 ± 0.50 | 1.15 ± 0.02 | 32.06 ± 0.46 | 3.35 ± 0.02 |
| Ox | 64.8 ± 0.35* | 1.29 ± 0.008* | 56.63 ± 0.75* | 4.46 ± 0.03* |
| anti-miR + ox | 77.65 ± 0.40** | 1.49 ± 0.017** | 75.06 ± 1.09** | 5.68 ± 0.02** |
| . | IL-6 (pg/mL) . | TNF-α (ng/mL) . | TGF-β (pg/mL) . | IL-2 (ng/mL) . |
|---|---|---|---|---|
| Mean ± SD | Mean ± SD | Mean ± SD | Mean ± SD | |
| Sham | 48.9 ± 0.80 | 1.0 ± 0.08 | 25.63 ± 0.91 | 2.8 ± 0.02 |
| anti-miR (25 nM) | 60.15 ± 0.35* | 1.075 ± 0.005* | 51.43 ± 0.64* | 4.29 ± 0.03* |
| anti-miR (50 nM) | 66.3 ± 0.52** | 1.23 ± 0.005** | 70.7 ± 0.95** | 5.09 ± 0.075** |
| Sham | 50.9 ± 0.50 | 1.15 ± 0.02 | 32.06 ± 0.46 | 3.35 ± 0.02 |
| Ox | 64.8 ± 0.35* | 1.29 ± 0.008* | 56.63 ± 0.75* | 4.46 ± 0.03* |
| anti-miR + ox | 77.65 ± 0.40** | 1.49 ± 0.017** | 75.06 ± 1.09** | 5.68 ± 0.02** |
Concentration of inflammatory cytokines secreted in 48 h culture supernatants by stimulated PMA-induced THP-1 cells from anti-miR (25 and 50 nM), ox-LDL and ox-LDL with anti-miR.
*P < 0.01 compared with control cell (sham).
**P < 0.01 compared with *group.
3.4 Analysis of ORP9 3′-UTR for miR-125a-5p responsiveness
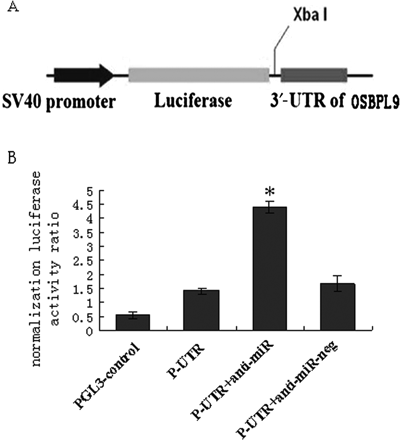
To find the target genes of the miR-125a-5p, we analysed candidates using the bioinformatics tools at multiple databases (targetscan4.1, PicTar, and miRanda). We surmised that ORP9 may be a target gene of miR-125a-5p. Therefore, the ORP9 3′-UTR was subcloned immediately down-stream from the firefly luciferase gene. THP-1 cells were cotransfected with miRNA-125a-5p inhibitor and ORP9-UTR, and relative luciferase activities were determined. The reporter gene assay showed that, compared with the pGL3-ORP9-3′‐UTR plasmid cotransfected cells, luciferase activity showed a 3.4-fold increased in the cells cotransfected with anti- miR-125a-5p inhibitor (Figure 4, P < 0.01). This indicated that miR-125a-5p directly targeted the ORP9.
Verification of the potential target genes of miR-125a-5p. (A) Schematic representation of the firefly luciferase (f-luc) reporter constructs utilized. (B) Cells were cotransfected with p-ORP9-UTR, pGL-3 control, and either negative control anti-miR (N.C.) or anti-miR-125a-5p at the concentration of 50 nM. Luciferase values were normalized by the Renilla control luciferase activity. The ratio of luciferase activity of each construct was calculated by luminometer. Data (mean ± SE) are from four independent experiments. *P < 0.01 compared with pGL3-ORP9-3′‐UTR plasmids only group.
3.5 MiRNA-125a-5p represses ORP9 transcript and protein expression in THP-1 cells
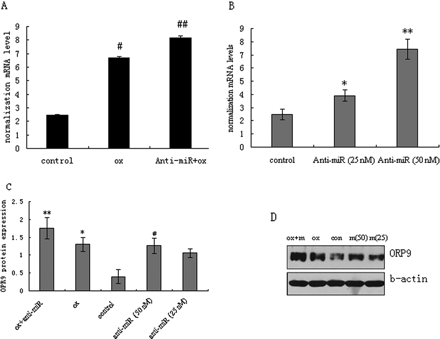
To verify the predictions and to study the functional consequences of miR-125a-5p, the THP-1 cell line was selected as a model for further study of the characteristics of miR-125a-5p regulation. In order to up-regulate ORP9 expression, we also use oxLDL as a stimulus. THP-1 cells were transfected with anti-miR-125a-5p and the human ORP9 mRNA and protein levels were quantified by TaqMan real-time PCR and western blotting. Anti-miR-125a-5p significantly induced ORP9 mRNA expression (Figure 5); the anti-miR (50 nM) group had a nearly three-fold increase compared with the control group (P < 0.01). However, the ORP9 mRNA level was lower in the oxLDL group than in the oxLDL plus miR-125a-5p inhibitor group (P < 0.05). The ORP9 protein expression consistently increased in the anti-miR group. Together, these results strongly suggested that ORP9 is one of the target genes of miR-125a-5p in monocytes.
Effects of miR-125a-5p inhibitor on ORP9 expression in oxLDL-stimulated THP-1 cells. Cells were induced with PMA for 24 h, and then transfected with miR-125a-5p inhibitor for 24 h followed by oxLDL stimulation. ORP9 mRNA and protein expressions were measured by real-time PCR and western blot. Representative real-time PCR and western blot results of ORP9 expression are shown. (A) ORP9 mRNA levels (with oxLDL treatment). (B) ORP9 mRNA levels (without oxLDL treatment). (C) Respective densitometric measurement results of ORP9. (D) ORP9 protein expression. The β-actin expression was used for protein level normalization. The band densities were measured by the Quantity One 1D analysis software program. Data (mean ± SD) were obtained from three independent experiments. Band density of native THP-1 cells were defined as control, **P < 0.01 compared with control; #P < 0.01 compared with *marked group; ##P < 0.01 compared with **marked group.
4. Discussion
Recently, several miRNAs have been found to be associated with macrophage inflammatory responses. Moschos et al.21 observed rapid and transient increase in miRNA expression in mouse lung following exposure to LPS and indicated this enhancement was correlated with proinflammatory cytokines. Pauley et al.22 found that peripheral blood mononuclear cells from patients with rheumatoid arthritis exhibit increases in miR-146a, miR-155, miR-132, and miR-16 expressions and they suggested that miR-146a function is essential for the regulation of TNF-α production. O'Connell et al.23 also found that miR-155 is substantially up-regulated in primary murine macrophages and is induced by a Toll-like receptor ligand, suggesting miR-155 as a common target of a broad range of inflammatory mediators. It is important to note that these previous data were obtained using either tumour cell (THP-1) models and human innate immune stimuli, or animal models, or rheumatoid arthritis patients. The limitations of these studies restrict extrapolation to what is occurring in monocyte/macrophage responses in atherosclerosis.
Therefore, considering that oxLDL is a major proinflammatory factor in the development of atherosclerosis, we chose oxLDL-stimulated human primary peripheral blood monocytes as a cell model to determine miRNAs associated with atherosclerosis. In the present study, we found, for the first time, that five miRNAs were aberrantly expressed in oxLDL-stimulated monocytes, with supporting results also obtained from TaqMan real-time PCR. These data, together with a global analysis of miRNA expression patterns, may allow us to identify several miRNAs involved in monocyte/macrophage responses to oxLDL stimulation and in the induction of atherosclerosis. Interestingly, our results differ from those of previous research obtained using a mouse lung model21 but are partially similar (miR-146a/b and miR-155) with those obtained with the THP-1 model.24,25 We believe this discrepancy is most likely ascribable to the differences between the in vitro and in vivo milieus and the differences between the stimuli. Additionally, our present study revealed, for the first time, that miR-125a-5p and miR-9 were also significantly up-regulated in monocytes after exposure to oxLDL.
In our present study, we found that miR-125a-5p was the most significantly up-regulated miRNA. We were able to predict the target mRNAs of miR-125a-5p using different algorithms, miRanda (http://microrna.sanger.ac.uk/), targetscan4.1 (http://www.targetscan.org/), and PicTar (http://pictar.bio.nyu.edu/). These algorithms may facilitate the identification of predicted target sites on the 3′-UTR of human gene transcripts for currently known mammalian miRNAs, based on interspecies conservation.16 We concluded, from the bioinformatics data, that ORP9 maybe the target gene of miR-125a-5p and found that the inhibition of miR-125a-5p increased the expression of ORP9 at both the mRNA and the protein levels. Furthermore, luciferase reporter assay analysis supported the supposition that the ORP9 gene is one of the direct targets of miR-125a-5p.
ORP9 belongs to the 12 member oxysterol binding protein (OSBP) family in humans, characterized by a conserved C-terminal OSBP homology domain that binds oxysterols, cholesterol, ergosterol, or phospholipids.26 Functional analysis of ORPs has indicated diverse roles in the regulation of lipid metabolism, which impacts on vesicle transport, cell cycle, and differentiation.27,28 As ORP9 is closely related to lipid metabolism and membrane transport,29,30 we therefore also explored the effects of miR-125a-5p on lipid uptake by oxLDL-stimulated macrophages. We demonstrated, by flow cytometry and by HPLC, that inhibition of miRNA-125a-5p expression significantly induced lipid uptake by macrophages. MiR-125a-5p could therefore decrease the lipid uptake in oxLDL-stimulated monocyte/macrophages. This activity may be the direct consequence of ORP-mediated lipid or sterol transport between membranes.
However, many studies have shown that the amount of lipid retained in macrophages also depends on the regulated uptake of oxidized lipoproteins by scavenger receptors, counterbalanced by degradation and efflux. We therefore measured the expression of scavenger receptors, CD68, LOX-1, and CD36. CD68/macrosialin, as a mucin-like scavenger receptor family protein, typically functions to clear cellular debris, to promote phagocytosis, and to mediate the recruitment and activation of macrophages.31 LOX-1 plays a fundamental role in the pathogenesis of atherosclerosis and contributes to plaque instability as well as to subsequent development of acute coronary syndromes.32 The importance of CD36 in atherogenesis has been recognized,33,34 although some recent studies have shown that the CD36 pathway does not ameliorate atherosclerosis35 and also triggers a proangiogenic signalling.36 Therefore, there is still controversy regarding the role of CD36 in atherogenesis. In our research, we measured the expressions of LOX-1, CD68, and CD36 and found that the expression of LOX-1 and CD68 were enhanced after inhibition by miRNA-125a-5p expression in oxLDL-stimulated macrophages, but amounts to no more than one-third of total oxLDL taken up by macrophages, and that CD36 expression showed no significant change. Our results indicate that miR-125a-5p can partly regulate oxLDL uptake of macrophages through reducing LOX-1 and CD68 expressions, but not via the CD36 pathway. The mechanisms for these effects of miR-125a-5p on the expression of LOX-1 and CD68 need further study.
ORP9 is also closely related to the adjustment of ERK phosphorylation and is relevant to the novel, independent predictors of coronary artery disease, namely ceramide and sphingomyelin synthesis.37, Additionally, previous research38 has indicated that miR-125 may be involved in the production of ROS-generating metal sulfates in neurocytes. MiR-125a-5p is up-regulated during neurotoxicity evoked by the rise of certain pro-inflammatory and pro-apoptosis genes.38 We therefore explored the effects of miR-125a-5p on the inflammatory response in oxLDL-stimulated monocyte/macrophages by ELISA assays. When compared with the cells incubated with oxLDL only, miR-125a-5p inhibitor significantly increased the secretion of IL-2, IL-6, TNF-α, and TGF-β. Previous studies have shown that IL-2, IL-6, and TNF-α are mainly pro-inflammatory cytokines of macrophages,2 although TGF-β has protective anti-inflammatory properties and blocks accelerated plaque formation.39–42 However, there is evidence that shows a pro-atherogenic role of TGF-β. In addition to its ability to promote fibrosis43,44 and neointima formation, TGF-β also has been found to markedly up-regulate the expression of high mobility group box 1,45 which can induce macrophages to increase in their atherosclerosis progress. All of these observations conflict with a protective role for TGF-β in atherosclerosis. Consequently, the role of TGF-β in the development of atherosclerosis needs further study.
Taken together, our results strongly indicate that miR-125a-5p may be an important regulator of the inflammatory response, lipid uptake, and ORP9 expression in oxLDL-stimulated monocyte/macrophages and that these may play a protective role against the development of atherosclerosis. We believe that once the functional role of miRNA has been further clarified, it will not be long before it can be used as a unique aid for fine-tuning atherosclerosis therapy targets.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Conflict of interest: none declared.
Funding
This work was supported by grants from the Fund of Xinhua Hospital, Shanghai Jiao Tong University School of Medicine (grant no. 2005-34) and the Science and Technology Commission Funding Project from Shanghai Municipality (grant no. 07JC14046).