-
PDF
- Split View
-
Views
-
Cite
Cite
Sander S.M. Rensen, Petra M.G. Niessen, Xiaochun Long, Pieter A. Doevendans, Joseph M. Miano, Guillaume J.J.M. van Eys, Contribution of serum response factor and myocardin to transcriptional regulation of smoothelins, Cardiovascular Research, Volume 70, Issue 1, April 2006, Pages 136–145, https://doi.org/10.1016/j.cardiores.2005.12.018
Close - Share Icon Share
Abstract
Objective Smoothelin-A and -B isoforms are highly restricted to contractile smooth muscle cells (SMCs). Serum response factor (SRF) and myocardin are essential for contractile SMC differentiation. We evaluated the contribution of SRF/myocardin to transcriptional regulation of smoothelins.
Methods Rat vascular SMCs were transfected with smoothelin-A and smoothelin-B promoter reporter constructs and promoter activity was analyzed. The effects of mutations in the smoothelin-A promoter CArG-boxes and co-transfections with a myocardin expression plasmid were assessed. Electrophoretic mobility shift assays and chromatin immunoprecipitations were performed to investigate SRF-binding to the smoothelin-A CArG-boxes.
Results Smoothelin promoter activity was detected in vascular SMCs. Comparative sequence analysis revealed two conserved CArG elements in the smoothelin-A promoter that bind SRF as shown by chromatin immunoprecipitation. The proximal CArG-near bound SRF stronger than CArG-far in gel shift assays. Mutagenesis studies also indicated that CArG-near is more important than CArG-far in regulating smoothelin-A promoter activity. Myocardin augmented smoothelin-A promoter activity 2.5-fold in a CArG-near-dependent manner. In contrast, myocardin had little effect on the smoothelin-B promoter.
Conclusion Smoothelin-A expression is controlled by an intragenic promoter whose activity is, in part, dependent on two CArG boxes that bind SRF. Our data show a role for SRF/myocardin in regulating smoothelin-A whereas the higher smoothelin-B expression appears to be SRF/myocardin-independent.
1. Introduction
Smooth muscle cells (SMCs) are of key importance for the correct functioning of the vasculature. To perform well, these cells are equipped with a unique set of proteins involved in their contraction and relaxation. Many of these proteins are exclusively or predominantly expressed in contractile SMCs, including α-smooth muscle actin (α-SMA), smooth muscle myosin heavy chain isoforms (SM-MHC), SM22α, smooth muscle calponin (SM-calponin), and smoothelin-A and -B [1,2]. During arterial remodeling, SMCs fulfil a role distinct from their contractile functions, which is aimed at restructuring or repairing to secure structural integrity. To accomplish this, they express a different set of proteins that allow proliferation, extracellular matrix (ECM) remodeling, and migration. Simultaneously, the expression of contractile proteins is downregulated. SMCs can shuttle between this so called synthetic phenotype and the contractile phenotype, a process referred to as phenotypic modulation [2]. During modulation towards the synthetic phenotype, expression of different contractile proteins does not diminish uniformly. Synthetic SMCs still express considerable amounts of α-SMA, SM22α, and SM-calponin. Expression of SM-MHC and smoothelins, however, is rapidly lost in modulating SMCs [3–7]. Therefore, SM-MHC and smoothelins are widely used markers of the contractile SMC phenotype.
In recent years, many studies have been performed to elucidate the mechanisms by which SMCs regulate the expression of proteins that are associated with the contractile phenotype. It has become clear that many cis-elements contribute to promoter activity of these genes, but one specific element, the CArG box, has been shown to be of critical importance for their expression [8–10]. This element, with the sequence CC(A/T6)GG, is recognized by the serum response factor (SRF), a transcription factor of the MCM1-Agamous-Deficiens-Serum response factor (MADS) box family [11]. Because SRF is widely expressed and not regulated in a SMC phenotype-specific way, it has long remained unclear how this factor would be able to direct expression of genes specifically in contractile SMCs. Recently, myocardin has been identified as an SRF cofactor with higher levels in contractile SMCs than in synthetic SMCs [12,13]. Myocardin is a member of the SAP (SAF-A/B, Acinus, PIAS) family of transcriptional regulators which strongly enhances transcription of most SMC contractile genes [13–16]. However, overexpression of myocardin in either embryonic stem cells or the SMC progenitor line A404 does not activate transcription of smoothelin-B [17], and there have been no studies on a possible activation of the smoothelin-A promoter by myocardin.
Expression of the smoothelin-A and -B transcripts has previously been shown to be mediated by two promoters separated by 10 kb in the human smoothelin gene [18]. The promoter upstream of exon 1 directs expression of smoothelin-B mRNA in vascular SMCs, whereas an intragenic promoter located upstream of exon 10 regulates smoothelin-A expression in both vascular and visceral SMCs. As with several other SMC marker gene promoters, CArG boxes are found in both human smoothelin promoter regions [19].
Since the presence of a promoter inside a gene's coding sequences is unusual, the first aim of the current study was to confirm the genuineness of the intragenic smoothelin-A promoter in the context of another species. Next, we set out to determine the functional importance of SRF binding to the two conserved CArG boxes that are present in the intragenic smoothelin-A promoter. The third aim was to assess the ability of myocardin to activate the smoothelin-A and smoothelin-B promoters in vascular SMCs.
We show that intragenic smoothelin-A promoter activity is partly dependent on CArG boxes, although their role is not as dominant as in other SMC-specific genes associated with the contractile phenotype. The activity of the smoothelin-B promoter is higher than smoothelin-A promoter activity, but not augmented by myocardin. Collectively, these findings suggest that distinct circuits of transcriptional control exist for smoothelin-A and smoothelin-B, and that non-CArG-dependent pathways predominate in the control of smoothelin expression.
2 Materials and methods
2.1 Cell culture
Pulmonary artery smooth muscle cells of the rat (PAC1) [20,21] were cultured in Dulbecco's modified essential medium supplemented with 10% fetal calf serum, 50 μg/ml gentamicin and 200 μM l-glutamine (Life technologies, Paisley, UK). The effect of low serum concentrations on smoothelin expression levels was determined by culturing PAC1 cells for three days in medium containing 5% or 2.5% serum.
2.2 Construction of plasmids
A mouse smoothelin-A promoter fragment was obtained by PCR amplification of a cosmid containing the mouse smoothelin gene [19], using the primers: FW 5′-GCCTTCCTTATGGAGGTGTG-3′, RV 5′-CCCAAGCTTCTGGCTCCCCAGTGCCAAC-3′, and Pfu Taq DNA polymerase (Stratagene, La Jolla, CA). PCR parameters were 94 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 58 °C for 30 s, 72 °C for 1 min, and a final step of 72 °C for 5 min. PCR products were cloned into pGEM-T Easy (Promega, Madison, WI) and sequence-verified. Promoter fragments were subcloned into the luciferase reporter vector pGL3-basic using restriction enzymes PvuII-HindIII (M-851 A) and XhoI-HindIII (M-1560 A), as indicated in Fig. 2A. These fragments extend to +58 bp relative to the smoothelin-A transcription start site [19]. Construction of α-SMA (− 195 bp) and human smoothelin-B promoter reporter plasmids (H-1616 B) has been described before [18,21].
A) Schematic representation of mouse smoothelin-A promoter constructs. The upper line represents the smoothelin gene structure, exons are indicated by white boxes and numbered. The bold arrow indicates the transcription start site of smoothelin-A, which resides inside exon 10 of smoothelin-B. The lower line shows the scale in bp. The CArG boxes are depicted as grey boxes. B) Comparison of activity of mouse smoothelin-A promoter fragments (M-1560 A and M-851 A) with human smoothelin-A (H-1613 A), human smoothelin-B (H-1616 B) and α-SMA promoters. Activity of the smoothelin-B construct is higher than activity of the smoothelin-A constructs. The active region in the mouse smoothelin-A promoter is located between − 1560 and − 851 nucleotides. Conserved CArG boxes are depicted as grey boxes, non-conserved CArG boxes as shaded grey boxes. The first exons of each gene are shown as white boxes.
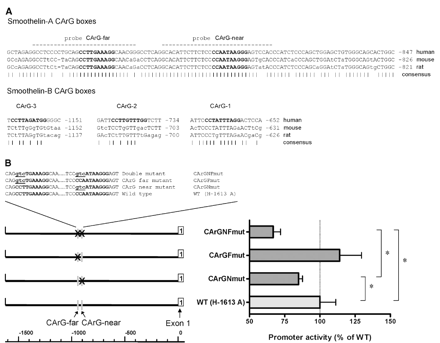
To determine whether the two CArG boxes of the human smoothelin-A promoter contribute to its transcriptional activity, they were mutated in the context of the reporter plasmid previously described to be active in PAC1 [18]. This H-1613 A plasmid was subjected to site-directed mutagenesis employing a high fidelity Pfu DNA polymerase (Stratagene). The proximal CC(A/T) sequence of each CArG box was mutated to GTC (see Fig. 3B), which is known to abrogate binding of SRF [9]. The resulting plasmids are referred to as CArGFmut, CArGNmut and CArGNFmut to indicate mutation of either the distal CArG-far box (Fmut), the proximal CArG-near box (Nmut), or mutation of both (NFmut). The mutated plasmids were verified by sequencing.
A) Sequence comparison of CArG box-containing regions in the smoothelin-A and smoothelin-B promoters of human, mouse and rat. Non-conserved nucleotides are indicated by small case letters. The CArG boxes are shown in bold. Dashes below the sequence indicate conserved nucleotides. Dashed lines indicate probes used for EMSA experiments. The numbers indicate the location of the sequence relative to the respective transcription start sites. B) Effect of CArG box mutations on the activity of the human smoothelin-A promoter construct H-1613 A. Mutation of the proximal CArG-near box caused a significant reduction in smoothelin-A promoter activity. Mutation of both CArG boxes simultaneously caused a further decrease. Statistically significant differences are indicated by an asterisk. The sequence of CArG mutations introduced into the human smoothelin-A promoter reporter construct are shown. CArG boxes are shown in bold, mutations are underlined and in small case.
2.3 Transient transfections and luciferase assays
1.5 × 105 PAC1 cells per well were seeded in 12-well plates and allowed to attach overnight. The next day, 2 μg of reporter plasmid were transfected into the 60–70% confluent cells using Fugene-6 transfection reagent (Roche, Brussels, Belgium) according to the provided instructions. A renilla reporter gene was included to normalize luciferase activity. To test the effects of myocardin expression on smoothelin-A and smoothelin-B promoter activity, co-transfection of a pCMV-Tag 2B vector carrying rat myocardin cDNA was performed. In these experiments, control cells were transfected with equivalent amounts of backbone vector without insert. After 48 h, analysis of luciferase expression in cell lysates was performed using a dual luciferase assay system (Promega) in a Biocounter M1500 luminometer (Lumac, Landgraaf, The Netherlands) or an AutoLumat LB 953 luminometer (EG and G Berthold, Gaithersburg, MD). All transfections were done in quadruplicate and repeated in three or more independent experiments.
2.4 Quantitative polymerase chain reaction (Q-PCR)
3 × 105 cells were seeded into 6-well plates and allowed to attach overnight. The next day, 1 μg of pCMV-Tag 2B vector carrying rat myocardin cDNA or 1 μg of empty vector was transfected into the cells using Fugene-6 transfection reagent (Roche) according to the provided instructions. Two days after transfection, total RNA was extracted using Tri reagent (Sigma-Aldrich, Zwijndrecht, The Netherlands) according to the manufacturer's instructions. Reverse transcription was performed using 1 μg of RNA in the iScript cDNA synthesis kit (Biorad, Hercules, CA). Quantitative real-time PCR was done using the MyIQ System (Biorad) and the following primers: SMTN/AB FW 5′-GGCTCGTCCACTCCAATG-3′, SMTN-A/B RV 5′-GGATGAGGAAGAGGAAGAGG-3′, SMTN-B FW 5′-TCGGAGTGCTGGTGAATAC-3′, SMTN-B RV 5′-CCCTGTTTCTCTTCCTCTGG-3′. Smoothelin mRNA levels were compared to α-SMA mRNA levels as determined by using the primer set α-SMA FW 5′-AACTGGTATTGTGCTGGACTCTGG-3′ and α-SMA RV 5′-CACGGACGATCTCACGCTCAG-3′. The cyclophilin transcript was used to normalize the amount and quality of the extracted RNA, using the following primer set: FW 5′-TTCCTCCTTTCACAGAATTATTCCA-3′, RV 5′-CCACCAGTGCCATTATGG-3′.
2.5 Nuclear extract preparation and electrophoretic mobility shift assays (EMSA)
PAC1 nuclear extracts were prepared from cells grown to confluency in 100 mm dishes using a Nuclear Extract kit (ActiveMotif, Rixensart, Belgium) and stored at − 80 °C. Protein concentrations were measured by the BCA assay (Pierce, Rockford, IL).
Probes for EMSA were obtained by end-labelling HPLC-purified oligonucleotides corresponding to the CArG boxes of the human smoothelin-A promoter (Sigma-Genosys, The Woodlands, TX). The oligonucleotides used are indicated in Fig. 3A (dashed lines), and extend from − 881 to − 911 (CArG-near) and − 919 to − 947 (CArG-far). In addition, the intronic CArG box (IC1) of the SM-calponin gene, which has previously been shown to strongly bind SRF [9], was used to compare SRF-binding affinities. 3.5 pmol of sense and antisense oligos were separately labelled using 10 μCi of [γ32P]ATP (at 4000 Ci/mmol) and T4 polynucleotide kinase (Promega). The labelled probes were mixed and annealed by slowly cooling to room temperature after heating to 70 °C. Unincorporated nucleotides and single-stranded oligos were removed by Sepharose G-50 spin columns (Amersham Biosciences, Piscataway, NJ). The labelling efficiency for the smoothelin probes was comparable, whereas labelling of the SM-calponin probe was 3-fold less. For competition experiments, unlabeled wild-type oligonucleotides or oligonucleotides mutated in the same way as the CArGFmut or CArGNmut reporter constructs (see Fig. 3B) were used at 20-fold excess.
Reaction mixtures for EMSA containing 5 μg of nuclear extract and 4% glycerol, 1 mM MgCl2, 0.5 mM EDTA, 0.5 mM DTT, 50 mM NaCl, 10 mM Tris–HCl (pH 7.5), and 0.05 mg/ml poly(dI–dC) were incubated for 10 min on ice. Subsequently, approximately 45000 cpm of labelled probe (15000 cpm for IC1) was added and incubated for an additional 20 min at room temperature. For supershifts, 1 μl of SRF antibody (Santa Cruz sc-335) was added to the indicated reactions followed by 15 min incubation at room temperature. The samples were loaded on a nondenaturing 4% 40:1 acrylamide/bisacrylamide gel which pre-ran for 15 min at 200 V prior to loading. Electrophoresis was carried out at 200 V for 25 min, after which gels were vacuum dried and exposed to X-ray film at − 80 °C.
2.6 Chromatin immunoprecipitation assay (ChIP)
ChIP assays were conducted with a kit according to the manufacturer's instructions (EZ ChIP; Upstate Cell Signaling Solutions, Lake Placid, NY). Briefly, PAC1 SMC were grown to subconfluency, fixed in 1% formaldehyde for 10 min to crosslink nucleoprotein complexes and scraped in phosphate buffered saline containing protease inhibitor cocktail. Pelleted cells were then lysed and sonicated 20 times at 10 s pulses using a Vibra Cell Sonicator (VC 130 PB; Sonics, Newton, CT) at 30% maximum output. Sheared DNA–protein complexes were immunoprecipitated with 2 μg anti-SRF (sc-335, Santa Cruz) or normal rabbit IgG as a control. Enriched DNA was then linearly amplified by PCR with primers flanking both CArG elements in the smoothelin-A promoter (FW primer, 5′-TGCCTACTCATCGGCAATTCTCCT-3′ and RV primer, 5′-TGGGTTCTGATGCTTCTCTAGCCT-3′).
2.7 Statistical analysis
All data are reported as mean±standard error of the mean and analyzed by one-way ANOVA, followed by Bonferroni's multiple comparison test. Values of P<0.05 are considered statistically significant.
3 Results
3.1 Smoothelin mRNA expression in PAC1 SMCs
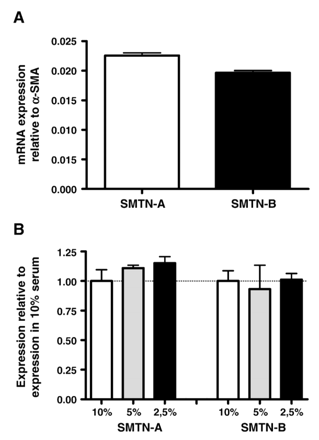
To investigate endogenous expression of smoothelins by PAC1 SMCs, we performed Q-PCR and compared expression levels of smoothelins to those of α-SMA. PAC1 cells expressed low levels of both smoothelin-A and smoothelin-B transcripts. Compared to α-SMA, the most abundant protein in SMCs comprising ∼40% of the total protein content, smoothelin-A levels were 44.3±0.9 times lower and smoothelin-B levels were 50.9±1.0 times lower (Fig. 1A). This shows that smoothelins are endogenously expressed in PAC1 cells and validates the use of these cells for functional analyses of smoothelin promoter activity.
A) Expression of smoothelin-A and smoothelin-B mRNA in PAC1 SMCs. Expression levels were assayed by Q-PCR and normalized to α-SMA expression. PAC1 cells express both smoothelin isoforms at the mRNA level. B) No significant effect of serum concentration on smoothelin-A and smoothelin-B mRNA expression in PAC1 cells. Expression levels were assayed by Q-PCR and normalized to expression at 10% serum.
Since serum concentrations have been reported to affect expression levels of smooth muscle specific markers, we examined the responsiveness of smoothelin transcript levels to reduced serum concentrations in the medium. Variations in serum concentrations ranging from 10% to 2.5% had no significant effect on the expression of either smoothelin-A or smoothelin-B, although there was a trend towards higher smoothelin-A expression with lower serum concentrations (Fig. 1B).
3.2 Functional analysis of the mouse smoothelin-A promoter
To confirm that the intragenic 5′ upstream region of the smoothelin-A transcription start site can activate transcription in SMCs, we constructed a reporter plasmid carrying 1560 bp of the putative mouse promoter (M-1560 A), and transfected it into PAC1 SMCs. A sixteen-fold induction of transcription compared to a promoterless construct was found, showing that this region, which is comparable to the previously tested human construct [18], indeed can act as a promoter in vitro (Fig. 2B). Interestingly, a smaller construct containing 851 bp of the promoter (M-851 A) was not able to drive transcription in PAC1 SMCs. Activities of a − 1616 bp human smoothelin-B promoter fragment (H-1616 B) and a α-SMA promoter fragment of 195 bp were approximately six-fold and three-fold higher in PAC1 cells compared to the previously tested human-1613 bp smoothelin-A promoter fragment (H-1613 A).
3.3 Functional significance of CArG boxes in the human smoothelin-A and -B promoter
Sequence comparisons between human, mouse and rat showed that the three CArG boxes in the human smoothelin-B promoter were not conserved across species (Genbank accession numbers: human AC005005; mouse AF132449; rat Ensembl gene ID: ENSRNOG00000019451). In contrast, the two CArG-boxes in the human smoothelin-A promoter and their surrounding sequences are highly conserved (Fig. 3A). A 111 bp region in intron 8 of the smoothelin gene containing the CArG boxes shows 86.5% homology. The 60 bp core of this region has only 7 mismatches between mouse, human and rat. The CArG boxes themselves are completely conserved, including a G substitution in the core of both CArG-far and CArG-near. The region between the CArG boxes, spanning 27 bp, shows only two non-conserved nucleotides between the three species. The 5′ flanking sequence of CArG-far is conserved for three nucleotides, like the 3′ flanking sequence of CArG-near. The high degree of homology indicated that the CArG boxes of the smoothelin-A promoter might be of functional significance.
To investigate the contribution of the CArG boxes to the transcriptional activity of the human smoothelin-A promoter, we introduced mutations in each element separately and in both elements simultaneously, and assessed activity of the mutated constructs in PAC1 cells (Fig. 3B). The introduced mutations are known to abolish SRF-binding to these cis-elements, rendering them inactive [9]. In comparison with the wild-type construct, mutation of the CArG-near box (CArGNmut) caused a mild but significant reduction of transcriptional activity of 15% (P=0.02). Mutation of CArG-far (CArGFmut) did not affect promoter activity significantly. In contrast, mutation of both CArG boxes simultaneously (CArGNFmut) reduced activity significantly by 32% compared to the wild type construct (P<0.01).
Surprisingly, co-transfection of wild type human smoothelin-A promoter fragments with constitutively active SRF (SRF-VP16) activated the promoter only slightly in PAC1 SMCs (statistically insignificant; data not shown). Similarly, human smoothelin-B promoter activity was not induced by constitutively active SRF.
3.4 SRF interaction with smoothelin-A promoter CArG boxes
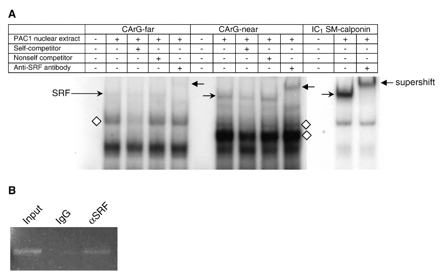
The relatively mild effect of the CArG box mutations on smoothelin-A promoter activity and the minimal activation of the promoter by SRF-VP16 led us to determine if SRF was able to bind to the smoothelin-A CArG boxes. To this end, we performed electrophoretic mobility shift assays (EMSAs) using PAC1 nuclear extracts and individual double stranded oligonucleotides containing either smoothelin-A CArG box and at least 6 bp of flanking sequence on each side. This revealed several specific nucleoprotein complexes with shifted mobility for both smoothelin-A probes (Fig. 4A), which were competed by unlabeled wild-type probe, but not by unlabeled mutant probe. The presence of SRF in one of these complexes was demonstrated by supershift experiments. SRF bound both smoothelin-A CArG probes, although binding to CArG-near was much stronger. However, in comparison to the IC1 CArG box of the SM-calponin gene which binds SRF strongly [9], even the smoothelin CArG-near bound SRF weakly. In addition, the IC1 CArG box experiment showed only one specific shifted band, representing the SRF-containing nucleoprotein complex. In contrast, both smoothelin-A CArG box regions bound additional proteins that are smaller than SRF and likely represent distinct transcription factors.
A) Electrophoretic mobility shift assay of smoothelin-A CArG-far and CArG-near boxes and of the intronic SM-calponin CArG box (IC1 SM-calponin). Both smoothelin-A CArG boxes bound several nucleoprotein complexes, including a SRF-containing complex. Supershifted complexes are indicated by a left pointing arrow, SRF bound to CArG boxes is indicated by a right pointing arrow, additional specific complexes binding the smoothelin-A CArG boxes are indicated by diamonds. B) ChIP assay results showing an enriched PCR product from PAC1 SMC nucleoprotein encompassing SRF and the tandem smoothelin-A CArG boxes following immunoprecipitation with antibody to SRF (αSRF). Control IgG shows little to no PCR product. The input lane represents genomic DNA prior to immunoprecipitation.
To determine whether SRF is bound to the smoothelin-A promoter CArGs in their native genomic context, we performed ChIP assays in PAC1 SMC. The results in Fig. 4B show enrichment for DNA containing the smoothelin-A promoter CArGs when nucleoprotein complexes are immunoprecipitated with SRF antibody. In contrast, little detectable DNA is seen when nucleoprotein complexes are immunoprecipitated with a control IgG. Note that because the CArG boxes are only 27 bp apart from one another, the ChIP assay cannot discriminate relative binding of SRF to each individual CArG element. Nevertheless, these data are congruent with EMSA and show that SRF binds to one or both of the smoothelin-A CArGs in vivo.
3.5 Activation of the smoothelin-A promoter by myocardin in a CArG-near dependent manner
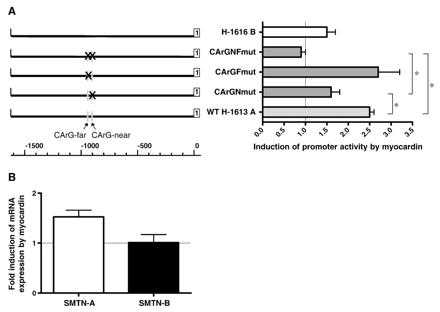
To assess the ability of myocardin to transactivate the smoothelin-A promoter, we performed transient reporter assays using either the wild-type promoter or the promoter with single or double CArG box mutations. Myocardin increased wild-type promoter activity 2.5-fold (Fig. 5A). Mutation of CArG-near greatly reduced the potential of myocardin to activate transcription (1.6-fold versus 2.5-fold induction). In contrast, mutation of CArG-far did not affect myocardin transactivation significantly (2.7-fold versus 2.5-fold induction). Upon mutation of both CArG boxes, the ability of myocardin to activate transcription was completely lost. Smoothelin-B promoter activity was augmented by myocardin, but to a lesser extent than smoothelin-A. Similar to previous reports, α-SMA promoter activity was highly stimulated by myocardin (17.1-fold induction).
A) Effect of myocardin overexpression on wild type (WT H-1613 A) and mutated (CArG–NF; CArG–F; CArG–N) human smoothelin-A and smoothelin-B (H-1616 B) promoter activity. The ability of myocardin to increase smoothelin-A promoter activity is reduced when SRF cannot bind CArG-near. Aberration of SRF-binding to both CArG-boxes causes a further decrease in promoter activity. Conserved CArG boxes are indicated by grey boxes. The first exons of each transcript are shown as white boxes. Statistically significant differences are indicated by an asterisk. B) Effect of myocardin overexpression on endogenous expression of smoothelins in PAC1 cells. Transfection with myocardin increased endogenous smoothelin-A expression, but did not affect smoothelin-B mRNA levels.
To determine if myocardin also increased endogenous expression of smoothelins in PAC1 cells, we transfected a myocardin expression plasmid into PAC1 SMCs and analyzed smoothelin mRNA levels by Q-PCR. In line with the results above, myocardin overexpression increased endogenous smoothelin-A expression, although the increase was small. In contrast, smoothelin-B mRNA levels did not increase after myocardin overexpression (Fig. 5B).
4 Discussion
SMC differentiation is monitored by marker genes whose expression is restricted to SMCs and representative of a given stage of differentiation. In recent years, it has become clear that studies on the regulation of marker gene expression can direct us to those cis-elements and trans-factors that are important for SMC differentiation itself. The recurring theme in this type of studies is the critical role of SRF-binding to one or more CArG elements in the promoters of SMC marker genes [11,22]. Many of these genes are downregulated when SMCs modulate towards a synthetic phenotype. The extent of downregulation is, however, quite variable between the different markers. The gene that seems to be most responsive to conditions promoting the synthetic phenotype is the smoothelin gene [4,23]. Elucidation of mechanisms that regulate the expression of this gene is therefore likely to contribute to the understanding of regulators of the contractile SMC phenotype.
Like several other genes whose expression is restricted to SMCs, the smoothelin gene generates several transcripts. Unlike these other genes, this is the result of a dual promoter system, instead of alternative splicing. An intragenic promoter fragment which resides in a region where several exons of smoothelin-B are located can drive transcription of the smaller smoothelin-A mRNA in humans [18]. Data presented in this study confirm and extend these results by showing that although smoothelin-B promoter activity was higher, a smoothelin-A promoter construct derived from the mouse smoothelin gene was consistently active in PAC1 SMCs. Of note, a Smad-binding cis-element in the first exon of the SM22α gene was recently demonstrated to contribute to SM22α expression [24], showing that exonic and regulating functions of the same sequence are not mutually exclusive.
The previously analyzed human smoothelin-A promoter fragment contained two conserved CArG elements in close proximity in an intronic region [19]. In this study, the functional significance of these elements has been tested. We show that the CArG boxes in the smoothelin-A promoter contribute to, but are not fully responsible for smoothelin-A promoter activity. This is in marked contrast to the promoters of other SMC restricted genes with paired CArG boxes like α-SMA and SM22α, which show 80% reduction of activity when either of their CArG boxes are mutated [25,26]. The discrepancy of these findings may be explained by the differential organization of the CArG elements between these genes, for CArG spacing and phasing have a major impact on promoter activity [27]. The distance between the CArG boxes in the α-SMA and SM22α genes is 40 and 113 bp, respectively, as compared to the smoothelin-A CArGs which are only 27 bp apart. This may lead to different binding potential of SRF or SRF-cofactors. Apart from this, particularly the smoothelin CArG-far has a relatively low affinity for SRF in EMSA experiments. CArG-near has a higher SRF-binding potential, which is in line with data on the affinity of SRF for CArG boxes with different G/C substitutions in the core indicating that a mutation at the last position of the AT core (CArG-near) is better tolerated than a mutation in the middle (CArG-far) [11]. Thus, it appears that CArG-near is the SRF-binding element that contributes most to smoothelin-A promoter activity. However, the additional reduction in promoter activity upon mutation of both CArG boxes compared to mutation of only CArG-near suggests that SRF binding to CArG-near may be required to facilitate SRF binding to CArG-far, as has been shown for the skeletal actin promoter [28].
In the gel shift experiments, additional nucleoprotein complexes with either smoothelin-A CArG box were formed that could not be supershifted by our SRF antibody. Since these complexes were smaller than the SRF-containing complex, this may indicate that distinct proteins unrelated to SRF bind these sequences. Alternatively, they may represent SRF–cofactor complexes of unknown nature, in which the cofactor masks the epitope recognized by our SRF antibody. Another possibility would be that an alternative isoform of SRF is involved [29], which is not recognized by the SRF antibody we employed. Regardless, the ChIP data show that SRF also binds one or both smoothelin-A CArG boxes in vivo, supporting the gel shift data and indicating a role for SRF in smoothelin-A promoter regulation.
Taken together, the results show that although SRF binding to the smoothelin-A CArG boxes is functional, this is not essential for smoothelin-A promoter activity. This suggests that the transcriptional activation of smoothelin-A, which is associated with the advanced contractile SMC phenotype, requires additional factors.
Several studies have documented the important roles of transforming growth factor-β (TGF-β) and various E-box binding transcription factors like GATA-factors and upstream stimulatory factors (USF)-1 and -2 in transcriptional regulation of other smooth muscle specific markers [2,24,30–33]. The active region in the mouse smoothelin-A promoter, which is located between − 1560 and − 851 nucleotides, contains three conserved putative USF-binding sites but no TGF-β control elements or Smad-binding elements [19]. Thus, further research may be directed towards a possible contribution of USF-1/USF-2 to regulation of smoothelin-A promoter activity.
In view of the recently suggested model in which myocardin specifically enhances transcription of genes containing two CArG boxes [16], we tested the ability of myocardin to increase transcription of smoothelin-A and -B. Smoothelin-B promoter activity was only marginally induced by myocardin, consistent with previous studies [17]. This is in line with the fact that the three CArG elements in the human smoothelin-B promoter have not been conserved across species. In contrast, myocardin did activate the smoothelin-A promoter, albeit to a much lesser extent than the α-SMA promoter. In agreement with the data on SRF binding, CArG-near but not CArG-far appeared to be important for smoothelin-A promoter activation by myocardin. Myocardin activity was completely lost when both CArGs were mutated, confirming that the smoothelin-A CArG boxes are functional. Similar results were obtained in a study of the α-SMA promoter, although mutation of only CArG-far severely impaired promoter activation by myocardin [15]. This suggests that activation of promoters in SMCs by myocardin is not absolutely dependent on two functional CArG motifs, but rather that one intact CArG element is sufficient. However, myocardin transactivation of the smoothelin-A promoter in PAC1 cells is much weaker than observed for other SMC genes [13], probably because the smoothelin-A CArG boxes do not bind SRF very efficiently. Thus, the hypothesized mechanism in which myocardin transactivational activity is only fully revealed after bridging of two CArG-bound SRF molecules is consistent with the present data [16].
It appears that other factors distinct from SRF or its coactivators are needed for full activation of the smoothelin promoters. This may explain the difference in the extent of downregulation of smoothelins versus other, SRF-dependent, contractile SMC genes in synthetic SMCs. Also, the possibility that the stronger upstream smoothelin-B promoter is required for full exploitation of the smoothelin-A promoter can not be ruled out, and would provide a novel contribution to the complex ways in which expression of SMC genes is regulated.
Acknowledgments
This work was supported by National Institutes of Health Grant HL-62572 (J.M.) and by grant D97.167 (S.R.) from the Netherlands Heart Foundation.
References
Author notes
Time for primary review 23 days
- polymerase chain reaction
- immunoprecipitation
- mutation
- plasmids
- serum response factor
- chromatin
- electrophoretic mobility shift assay
- gel
- genes
- muscle contraction
- mutagenesis
- protein isoforms
- sequence analysis
- transfection
- mice
- rats
- transcriptional control
- binding (molecular function)
- myocytes, smooth muscle