-
PDF
- Split View
-
Views
-
Cite
Cite
Carmen López-Sánchez, Virginio García-Martínez, Molecular determinants of cardiac specification, Cardiovascular Research, Volume 91, Issue 2, 15 July 2011, Pages 185–195, https://doi.org/10.1093/cvr/cvr127
Close - Share Icon Share
Abstract
In this review, we report and analyse the molecular factors involved in cardiogenesis from the earliest stages of development, using mainly the chick embryo as a model. The first part of the review demonstrates the areas where cardiogenic cells are located from gastrula stages, analysing a brief summary of the fate map of cardiogenic cells, from the epiblast through to the primitive heart tube. The next part analyses the commitment of pre-cardiac cells in cardiogenesis before, during, and after ingression through the primitive streak. Throughout the different journeys of the pre-cardiac cells, from the origin on the epiblast level up to the constitution of the tubular heart in the mid-line, the genes involved in the different stages of the process of cardiogenesis are very numerous. These have a greater or lesser importance depending on their specificity and the order in which they appear, bearing in mind that they become more valuable as the developmental process advances and the precursor cells start acquiring the commitment of pre-cardiac cells. Next, we show some box-filled diagrams to illustrate the dynamic gene expression pattern throughout the early stages of heart development, grouping the genes by their chronological significance. Finally, we discuss the implications that this temporal genomic expression could have in the induction and specification of the different types of cells and regions of the heart.
1. Introduction
Over the last few years, the challenge to understand the molecular determinants responsible for heart development has taken on special relevance. Here, we analyse the molecular determinants of cardiac specification of the cells programmed to form the heart. Thus, we will analyse the origin, location, and migration of the prospective cardiogenic cells as well as the possible commitment of these groups of cells to form cardiac or other structures, their molecular characteristics, specific gene expression patterns, and finally the inductive or repressive factors involved in cardiac specification.
We will use the chick embryo as the main model to describe cardiac development, and we will make specific comments about other species and models, mainly the mouse, when relevant information is available. The number of genes implicated is very large, but no synthesis has brought the information together in a comprehensive manner. Here we use a box-filled diagram to show the dynamic gene expression pattern throughout the early stages of development, grouping the genes not for their proteomic or genomic lineage as is usual in other works, but by their chronological significance in cardiogenesis. We then discuss the implications that this temporal genomic expression could have in the induction and specification of the different types of cells and regions of the heart.
2. Origin, location and migration of prospective cardiogenic cells
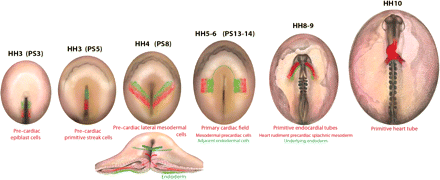
In avian embryos at stages 3a and 3b (Hamburger and Hamilton;1 substaging of stage 3 as described by Schoenwolf et al.2) and PS2–PS3 (as described by Lopez-Sanchez et al.3), the prospective cardiogenic cells are located in the epiblast and primitive streak (Figure 1). Within the epiblast, these cells are bilaterally distributed on both sides of the primitive streak, caudal to the node.4,5 In the mouse, the origin and position of the epiblast prospective cardiogenic cells are similar to the chick.6,7 In the zebrafish, the cardiac precursors are in the ventral marginal zone at the mid- to late blastula stages.8,9
Schematic diagrams showing the pre-cardiac cells, from stage HH3 (PS3) to stage HH10, in the developing chick embryo. Red indicates the mesodermal pre-cardiac cells, and green the endodermal cells that are related to cardiogenesis. The section shows the migration of epiblast cells ingressing through the primitive streak at stage HH4.
A few hours later (PS5), the rostral half of the primitive streak, with the exception of the node, contains the prospective cardiogenic cells10 (Figure 1), and also the cells that are going to form the endoderm which underlies the pre-cardiac mesodermal cells at both sides of the embryo.5,11,12 Progeny of cells from this region will contribute to all layers of the heart tube, including endocardium, myocardium, and parietal pericardium.2,10 A similar localization and fate of cardiogenic cells have been described in the mouse.13 However, it is still not clear whether the co-alignment of cells in the primitive streak and heart tube, and the endocardial and myocardial progenitors in the primitive streak, represent completely separate lineages, as has been found in the chick, is also true for the mouse.14
Later (HH4) in gastrulation (Figure 1), pre-cardiac cells from the epiblast invaginate through the primitive streak, and move bilaterally and cranially,15 to form the bilateral cardiogenic mesoderm,16 located in the anterior position, and constituting the primary cardiac field, also called the heart-forming region. At this moment, the pre-cardiac mesoderm is surrounded by the adjacent endoderm, which comes from the more cranial part of the primitive streak, and which plays a crucial role during cardiac specification.12
The primary cardiac field does not show precise boundaries, and the discussion about the limit is still open.17,18 Some studies even described a single cardiogenic field as a ‘horseshoe-shaped zone’ or ‘cardiac crescent’ continuous across the mid-line, cranial to the pre-chordal plate.19,20 The organization of the heart-forming region in two clearly separate regions, as described in the chick embryo at stage HH5, with a gap between them, has also been shown in amphibians. In the mouse, it is difficult to see independent bilateral cardiogenic fields, and they are usually described as a single, crescent-shaped region.21–23
From stage HH7 begins the organization of the heart rudiment pre-cardiac splachnic mesoderm, which will form both of the primitive endocardial tubes, surrounded by the underlying endoderm. From stage HH9, both primitive endocardial tubes fuse in the mid-line to form the primitive heart tube, structurally organized in concentric layers of endocardium and myocardium.24
2.1 Commitment of cardiogenic cells
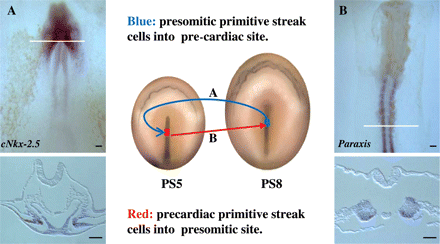
The fate maps clearly show that the cells which give rise to the heart come from the epiblast layer, primitive streak, and lateral mesoderm, and lead us to question at what point during this process the cells commit to differentiate towards cardiac specification. Numerous experiments have been conducted in order to answer this question, mainly based on the transplantation of cardiogenic cells to areas that are not involved in the formation of the heart, as well as the contrary; for example, there are experiments based on the grafting of prospective primitive streak cardiac cells into the prospective somitic cells, and vice versa12,25,26 (Figure 2), grafting of prospective primitive streak cardiac cells into the germ cell crescent,12 rotation of craniolateral endoderm,26 rotation of mesodermal pre-cardiac cells and the underlying endoderm,27 or explants of cardiogenic mesoderm, with or without endoderm.11,28,29 Experimental studies in the mouse have shown that the epiblast-derived cells differentiated into myocardial cells, after being transplanted directly to the cardiogenic field of the late primitive streak embryo without ingression through the primitive streak or moving within the mesoderm, suggesting that ingression is not critical for specification of myocardial fate.30,31
Schematic diagram showing the experimental procedure for the reciprocal transplantation (quail–chick chimera) of primitive streak cells, between embryos at stages PS5 (pre-cardiac cells, in red) and PS8 (pre-somitic cells, in blue). (A) Whole mount and section after in situ hybridization and immunocytochemistry (anti-quail) showing the quail pre-somitic cells (brown), transplanted into the pre-cardiac site, expressing cNkx-2.5 at the level of the pre-cardiac splanchnic mesoderm. (B) Whole mount and section after in situ hybridization and immunocytochemistry showing the quail pre-cardiac cells (brown), transplanted into the pre-somitic site, expressing Paraxis (specific somitic marker) at the level of the somites. The white line indicates the level of the respective section. Scale bars represent 400 µm.
3. Spatial and temporal gene expression that correlates with cardiac specification
In this section, we provide a detailed temporal and spatial analysis of the genes that are expressed in the cellular groups which are going to contribute to the formation of the tubular heart, with special reference to data obtained through in situ hybridization in avian embryos.
3.1 Gene expression patterns during gastrulation
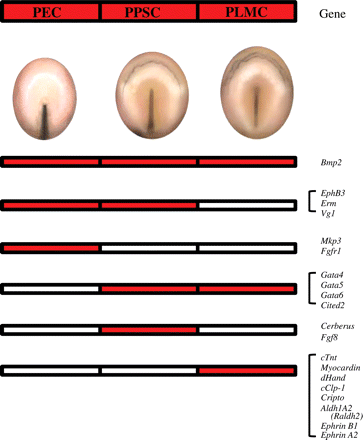
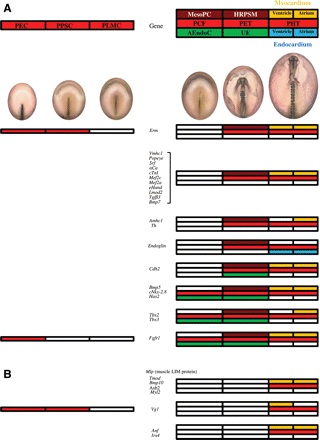
Several genes are expressed from the initial stages of cardiogenesis. Figure 3 illustrates in red boxes the expression of different genes in pre-cardiac cells according to their location at the level of the epiblast, primitive streak, and lateral mesoderm. Bmp232 is expressed in pre-cardiac cells, from stage PS3 in the epiblast, continuing at stage PS5 in the primitive streak, and at stage PS8 in the pre-cardiac lateral mesoderm. In contrast, EphB3 (Eph receptor tyrosine kinase33,34), Erm (transcriptional targets of Fgf signalling35,36), and Vg1 (a member of the Tgfβ superfamily37,38) are expressed only in the pre-cardiac cells of the epiblast and the primitive streak, but not in pre-cardiac lateral mesodermal cells. Moreover, Mkp3 (a MAP kinase phosphatase 336) and Fgfr136,39 are expressed only in pre-cardiac epiblast cells. Some other genes, Gata-4, -5, and -6 (zinc finger proteins40,41), and Cited2 (formerly melanocyte-specific gene related gene, MRG142), are expressed in pre-cardiac cells of the primitive streak and lateral mesoderm, whereas Cerberus (secreted Bmp and Wnt antagonist43,44) and Fgf838 are expressed only in pre-cardiac primitive streak cells. Finally, cTnt (cardiac troponin T, responsible for binding of the troponin complex to tropomyosin45,46), Myocardin (a serum response factor cofactor47), dHand (member of a family of muscle-specific basic helix–loop–helix ‘bHLH’ transcription factors48,49), cClp-1 (cardiac lineage associated protein, related to Mef250), Cripto (formerly Tdgf1, teratocarcinoma-derived growth factor51,52), Aldh1A2 (Raldh2; retinaldehyde deshydrogenases, related to retinoic acid53,54), EphrinB1, and EphrinA2 (both ligands of the Eph family of receptor tyrosine kinases34) start to be expressed from pre-cardiac lateral mesodermal cells. These genes are also expressed later during cardiac specification and differentiation, with different roles.
The diagrams show in red the expression of different genes in pre-cardiac cells according to their location, as follows: pre-cardiac epiblast cells (PEC), pre-cardiac primitive streak cells (PPSC), and pre-cardiac lateral mesodermal cells (PLMC).
3.2 Gene expression patterns of ingressed pre-cardiac mesodermal cells
The primary cardiac field, constituted by the mesodermal pre-cardiac cells at stage HH5 (PS13), does not show a precise boundary or limit. There has been an attempt to establish a correlation between the pattern of expression of some genes with the limits of the primary cardiac field. cNkx-2.5 has been proposed as the first transcription factor responsible for the beginning of cardiogenesis; nevertheless, its expression does not coincide exactly with the described cardiac fate map.17,18,28 An attempt has also been made to relate these limits to the expression of Bmp2,32 but in the same way a precise correlation with the fate maps does not exist.18
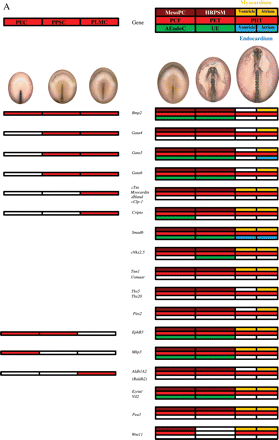
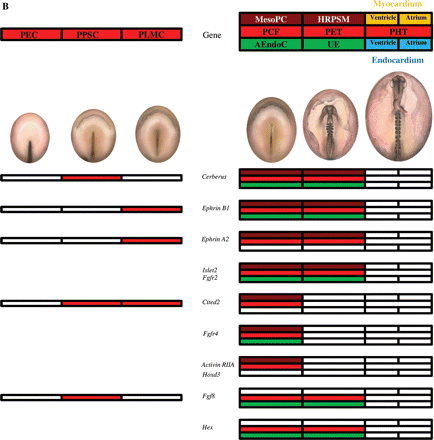
Figures 4 and 5 illustrate in red boxes the expression of different genes in cardiogenic cells according to their location at the level of the primary cardiac field, primitive endocardial tubes, and primitive heart tube. Moreover, brown and green boxes indicate the expression at the level of the mesoderm and/or endoderm, respectively.
Genes expressing from HH5, in mesodermal pre-cardiac cells (MesoPC), reaching the primitive heart tube (A) or not (B). The diagrams on the left show in red the expression of different genes as described for Figure 3, but in this case the expression of the same genes can be also detected later in development, as indicated on the right. The expression of the different genes at the level of the primary cardiac field (PCF), primitive endocardial tubes (PET), and primitive heart tube (PHT) is indicated by colors. When a gene is specific for the primary cardiac field, it may be expressed at the level of the mesodermal pre-cardiac cells (MesoPC, in brown) and/or the adjacent endodermal cells (AEndoC, in green). When a gene is specific for the primitive endocardial tube, it may be expressed at the level of the heart rudiment pre-cardiac splanchnic mesoderm (HRPSM, in brown) and/or the underlying endoderm (UE, in green). Finally, the genes expressed at the level of the primitive heart tube may be located in the myocardium (in yellow) and/or endocardium (in blue), corresponding to the ventricle (half left side) or atrium (half right side).
(A) The genes expressed from HH8, in heart rudiment pre-cardiac splachnic mesoderm (HRPSM) reaching to the primitive heart tube. (B) The genes expressed from stage HH10, at the level of the primitive heart tube. The diagrams on the left show in red the expression of different genes as described for Figure 3, but in this case the expression of the same genes can also be detected later in development as indicated on the right. The expression of the different genes at the level of the primary cardiac field (PCF), primitive endocardial tubes (PET), and primitive heart tube (PHT) is indicated by colors. When a gene is specific for the primary cardiac field, it may be expressed at the level of the mesodermal pre-cardiac cells (MesoPC, in brown) and/or the adjacent endodermal cells (AEndoC, in green). When a gene is specific of the primitive endocardial tube, it may be expressed at the level of the heart rudiment pre-cardiac splanchnic mesoderm (HRPSM, in brown) and/or the underlying endoderm (UE, in green). Finally, the genes expressed at the level of the primitive heart tube may be located in the myocardium (in yellow) and/or endocardium (in blue), corresponding to the ventricle (half left side) or atrium (half right side).
Gata-4, -5, -6,42,55cTnt,46Myocardin,47dHand,48,49cClp-1,50 and Cripto,52 are expressed at the level of the mesodermal pre-cardiac cells, as well as in pre-cardiac cells in earlier stages. However, Smad6 (a putative negative regulator of Bmp, Tgfβ, and activin signalling56–58), Tnn1 (troponin I type 149), Usmaar (smooth muscle α-actin23), Tbx5, Tbx20 (T-box transcription factors59–62), and Pitx2 (a member of the bicoid-related family of homeobox-containing genes63) are not expressed in pre-cardiac cells in earlier stages. In contrast, EphB3,33,34Mkp3,36Aldh1A2 (Raldh2),53,54Ezrin (Vil2; a member of the ezrin–radixin–moesin family of actin binding proteins64), Pea3 (transcriptional targets of Fgf signalling35,36), and Wnt1165 are less specific to cardiac specification, because they are also expressed in the neural plate, paraxial mesoderm, or caudal hindbrain (Figure 4A). All these genes continue to be expressed at the level of the primitive endocardial tubes (with the exception of Wnt11) and primitive heart tube. Several other genes are excluded from this rule, as follows (Figure 4B): Cerberus,43,49EphrinB1, and EphrinA2,34Islet2 (a member of a family of homeodomain-containing transcription factors that possess an amino-terminal pair of zinc-binding LIM domains49,66), and Fgfr2.36 Moreover, Cited2,42Fgfr4,36Activin RIIA (with ability to bind activin-A67), and Hoxd3 (a retinoic acid-dependent Hox gene68–70) are limited to the primary cardiac field. Furthermore, Fgf8 (which is expressed in pre-cardiac primitive streak cells;38,71Figure 6A) and Hex (a homeobox gene also known as Prh72) are not even expressed in heart rudiment pre-cardiac splachnic mesoderm, but only in the underlying endoderm.
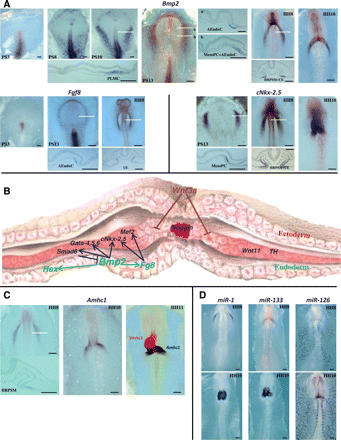
(A) Whole mount in situ hybridization of chick embryos from stage PS3 to HH10 showing the expression of three genes related to cardiogenesis: Bmp2, Fgf8, and cNkx-2.5. Sections of some embryos (indicated by white lines) reveal the expression in pre-cardiac lateral mesodermal cells (PLMC), mesodermal pre-cardiac cells (MesoPC), adjacent endodermal cells (AEndoC), heart rudiment pre-cardiac splachnic mesoderm (HRPSM), and underlying endoderm (UE). Scale bars represent 400 µm. (B) Schematic model illustrating the molecular determinants of cardiac specification. Endoderm-derived signalling molecules, Bmp2 and Fgf8, are involved in the induction of cardiac mesoderm, inducing the expression of cNk-2.5, Gata-4, Gata-5, Gata-6, and Mef2. Smad6 (a negative regulator of Bmp signalling) expression is induced by Bmp2, which also regulates Fgf8 and Hex. Wnt11 and the tyrosine hydroxylase gene (Th) are also expressed in cardiac mesoderm. Antagonists from the ectoderm, Wnt3a, and secreted from the notochord, noggin, suppress cardiac specification. (C) Whole mount in situ hybridization of chick embryos showing the expression of atrial (Amhc1) and ventricular (Vmhc1) markers. Amhc1 is illustrated at the level of the primitive endocardial tubes (HH8) and primitive heart tube (HH10). White line indicates the level of the section showing the expression in the heart rudiment pre-cardiac splachnic mesoderm (HRPSM). The third panel shows a double in situ hybridization (HH11) showing the regionalization during cardiac looping. Amhc1 (dark blue) and Vmhc1 (red). Scale bars represent 400 µm. (D) Whole mount in situ hybridization of chick embryos showing the expression of micro-RNAs (miR-1, miR-133, and miR-126) at the level of the primitive endocardial tubes (HH8, top panels) and primitive heart tube (HH10, bottom panels). Scale bars represent 400 µm.
Some of the genes previously referred to are also expressed at the level of the adjacent endodermal cells of the primary cardiac field and/or primitive endocardial tubes (Figure 4), as follows: Bmp2 (Figure 6A), Gata-5, Gata-6, Smad6, cNkx-2.5 (Figure 6A), EphB3, Mkp3, Ezrin (Vil2), Cerberus, EphrinB1, Islet2, Fgfr2, and Fgfr4; these probably play some relevant role in the regulation of mechanisms of cardiogenesis.
The following group of genes (Figure 5A) begins to express at the level of the heart rudiment pre-cardiac splanchnic mesoderm during the formation of the primitive endocardial tubes: Erm,36Vmhc1 (ventricular myosin heavy chain73), Popeye (Popdc2; encoding protein of the plasma membrane in muscle cells74), Srf (serum response factor, a member of the MADS-box75), αCa (cardiac α-actin75), cTnI (cardiac troponin I76), Mef2c (muscle enhancer factor 2 protein71), Mef2a,77eHand (member of a family of muscle-specific basic helix–loop–helix ‘bHLH’ transcription factors48), Lmod2 (leiomodin, an actin nucleator in cardiomyocytes49,78), Tgfβ3,49Bmp7,76Amhc1 (atrial myosin heavy chain69), Th (tyrosine hydroxylase79), Endoglin (a co-receptor for the Tgfβ superfamily80,81), Cdh2 (cadherin 2 type 149), Bmp5,76cNkx-2.8 (a member of the Nk-2-family of transcription factors82), Has2 (hyaluronan synthase enzyme83), Tbx2, and Tbx3.84 Immunohistochemistry revealed Fgfr1 at the level of the endoderm in the primary cardiac field, followed by localization in the pre-cardiac splachnic mesoderm and underlying endoderm at stage HH8, and becoming confined to the myocardium of primitive heart tube.85–87 The additional expression in the underlying endoderm of Cdh2, Bmp5, cNkx-2.8, Has2, Tbx2, Tbx3, and Fgfr1 probably plays a relevant role in the regulation of mechanisms of cardiogenesis.
3.3 Gene expression patterns of primitive heart tube formation
During the formation of the primitive endocardial tubes (HH8) and primitive heart tube (HH10), the expression of a great number of genes listed in Figures 4A and 5A is maintained. Vmhc1 is progressively restricted to the ventricular (anterior) sector, possibly regulated by the expression of Irx4. Vg1 and Anf also adopt a pattern of expression restricted to the anterior (ventricular) sector of the tube, suggesting a role in ventricular differentiation. In addition, the atrial (posterior) sector is regulated by the expression of Amhc1, the first marker described for atrial specification. Now we know that the commitment to atrial differentiation, as well as its caudal prolongation towards the venous pole, is regulated by Th expression.79 Moreover, other molecular factors (Figures 4A and 5A) have also been proposed as determinants of atrial specification, as follows: Tbx2, Tbx3, Aldh1A2 (Raldh2), Tbx5, Tbx20, Gata-4, -5, -6, and Bmp2. Special reference must be given to Gata-5,55 and to Smad6 (possibly regulated by Bmp signaling;58Figure 4A) and Endoglin81 (Figure 5A), which are expressed during differentiation of endocardium.
To conclude the description of the pattern of gene expression, we will finish with the genes (Figure 5B) that start to be expressed at the level of the primitive heart tube, as follows: Mlp (muscle LIM protein, also called Csrp3, cysteine and glycine-rich protein 349), Tmod (tropomodulin 149), Bmp10,76Asb2 [ankyrin repeat and suppressor of cytokines signalling (SOCS) box protein 249], Myl2 (myosin regulatory light chain 2A49), Vg1,88Anf (atrial natriuretic factor89), and Irx4 (iroquois-related homeobox gene90).
4. Molecular regulation of cardiac specification
It has been proposed15 that the expression of Wnt3a at the level of the primitive streak acts as a chemo-repellent signal to guide the movement of cardiac progenitor cells away from the streak, resulting in lateral migration.
Although ingression of cardiogenic mesoderm is very similar to that in the chick, in the mouse a mesodermal marker, Mesp1, was used to trace the first pre-cardiac mesodermal cells that ingress through the primitive streak,91 suggesting that migration of the cells from the primitive streak to form the cardiogenic field in the mouse depends on expression of this gene.92
We have previously reported12,93 that transplanted Hensen's node from quail donor to pre-cardiac and non-pre-cardiac regions of chick host embryos is able induce specific cardiac markers, suggesting that this organizer could participate in cardiac specification, probably by means of secreted growth factors (Fgf8 and Bmp2), which are being expressed in Hensen's node (Figure 6A).
It is assumed that specification of the cardiac fate requires close association between the mesoderm of the primary cardiac field and the underlying endoderm. This model proposes that cNkx2.5 expression in the primary cardiac field is induced by Bmp2, emanating from the adjacent endoderm28,94 and is based on misexpression experiments in chick embryos in which ectopic exogenous application of Bmp2 to the tissue medial to the primary cardiac field resulted in induction of cNkx2.5 expression.32,94 Furthermore, exogenous Bmp2 is also capable of inducing the expression of Fgf8,71Gata-4,32,94Gata-5, Gata-6,42Hex,95 and Smad6,58 as well as Tbx2 and Tbx3, all of which are related to cardiogenesis.84 Also, incubation of pre-cardiac mesoderm with noggin, an antagonist of Bmp signalling,58,96 inhibits cardiac myogenesis and expression of myocardial marker genes (cNkx-2.5, Gata-4, Mef2a, eHand, and Vmhc142).
It has also been shown that several members of the Fgf family are capable of inducing cardiogenic markers in non-pre-cardiogenic mesoderm. Thus, the administration of Fgf8 to the tissue lateral to the primary cardiac field results in cNkx2.5 (and Mef2c), but not Bmp2 and Gata-4, induction,71 which are expressed from the time of early specification of cardiac precursor cells. Moreover, we have shown previously93 that Fgf2 and Fgf4 are able to induce the expression of cNkx2.5 and cNkx2.8 in non-pre-cardiogenic tissue, at the level of the germinal cell crescent. Interestingly, in some other non-pre-cardiogenic tissues, such as explanted caudal lateral mesoderm, a combination of Bmp2 and Fgf4, but neither factor alone, is able to induce cNkx-2.5 expression.97,98 Administration of Fgf1, 2 or 4 (which are not expressed in pre-cardiac cells) to cultured pre-cardiac mesoderm cells99 resulted in the formation of vesicles containing an adherent multilayer of synchronously contractile cells, supporting their participation in cardiogenesis and subsequent development of the embryonic myocardium. All these data suggest that initial cardiac specification is regulated by complex relationships between the Bmp and Fgf signalling pathways (Figure 6B).
Wnt11, which is expressed in early avian mesoderm in a pattern that overlaps with the pre-cardiac regions,65,100,101 can promote cardiac development within non-cardiac tissue, suggesting that it may also play a role in the formation of the vertebrate heart.102–104 In contrast, at the level of the posterior lateral mesoderm, Wnt3a and Wnt8c signalling act to inhibit cardiac differentiation.105
Here we show a group of genes being expressed specifically at the level of this underlying endoderm in relation with the primary cardiac field, as follows: Fgf8 and Hex (Figure 4B; also Fgf1, 2 and 4, identified by immunohistochemistry99), as well as Bmp5, cNkx-2.8, Has2 (which is independent of Bmp2 signalling83), Tbx2, Tbx3, and Fgfr1 (Figure 5A). In a second group of genes, we include those that are expressed not only in the adjacent endodermal cells (HH5), but also at the level of the pre-cardiac mesoderm, strongly related with cardiac specification (Figure 4), as follows: Bmp2, Gata-5, Gata-6, Cripto, Smad6, EphB3, Mkp3, Ezrin, Cerberus, EphrinB1, Islet2, Fgfr2, and Fgfr4. A third group of genes is also expressed in the underlying endoderm of the heart rudiment pre-cardiac splachnic mesoderm, and start to be expressed from stage HH8, as follows: cNkx-2.5 and Cdh2. Thus, there may be different signals for sequential stages that may be needed for the initial steps in the establishment of the fate of cardiac cells and others in the later steps of cardiac cell-specific differentiation.
Some genes that were initially expressed throughout the whole myocardium become restricted to the presumptive atria and ventricular segments (Figure 6C). Vmhc1 is expressed in the heart rudiment pre-cardiac splachnic mesoderm and primitive heart tube in all myogenic populations, but its expression is later restricted to ventricular myocytes.73 The posterior heart segment expresses Amhc1, and it develops into the atria.69 The early activation of Amhc1 expression in the posterior cardiac myocytes suggests that the anterior heart progenitors differ from the posterior heart progenitors in their myosin isoform gene expression. It has been proposed that in the chick embryo, Irx4 (which is downstream of cNkx-2.5 and dHand, and expressed exclusively in the ventricle106) regulates the chamber-specific expression of myosin isoforms by activating Vmhc1 and suppressing Amhc1 in the ventricle, while morphogenesis is apparently not affected by this homeobox gene.90,106
The transcription factor genes Hand1 and Hand2 are also co-expressed in the primitive heart tube, but later they accumulate in the ventricular segment.48,107,108 Some other transcription factors have been identified and, perhaps, are involved in establishing anterior–posterior patterning of the primitive heart tube. For example, Hey1 and Hey2 are expressed in atrial and ventricular progenitor cells, respectively.109,110
Furthermore, in the chick and the mouse, the atrial natriuretic factor (Anf) gene is initially expressed throughout the whole myocardium of the primitive heart tube, and becomes restricted to the ventricular segment.89 In contrast, Gata-4 and Tbx5, with expression throughout the primary cardiac field, become restricted to the atrial compartment.40,84,111 It has been proposed that Anf is regulated by a co-operation of Gata-4 and Tbx5, as well as cNkx-2.5, which are expressed earlier in cardiogenesis.62,112 However, Tbx20 (which is induced by Bmp2) represses Anf promoter activity and also inhibits the activation mediated by Tbx5.62 Our own recent results add more information about the mechanism to generate atrio-ventricular specific gene expression. We revealed79 a novel function of Th (tyrosine hydroxylase) in cardiac development, acting in concert with additional factors to define multiple aspects of atrial identity. Th expression, localized to the heart rudiment pre-cardiac splachnic mesoderm, induces Amhc1 and Tbx5, and suppresses Irx4 and Vmhc1. The relationship between Gata-4 expression and retinoic acid effects has been previously reported.113,114 Furthermore, the gene for the retinoic acid-synthesizing enzyme, Raldh2, is specifically expressed within the atrial region.115 Interestingly, Th might be a putative downstream target of retinoic acid activity in establishing the anterior–posterior heart tube axis.79
Recently, the identification of micro-RNAs expressed in specific cardiac cell types has led to the discovery of important regulatory roles for these small RNAs during cardiomyocyte differentiation and cell proliferation. In the chick and the mouse, the expression of miR-1 and miR-133 has been described during cardiac development.116,117 As can be seen in Figure 6D, miR-1 and miR-133 are expressed in pre-cardiac mesoderm, and later expressed in the myocardium of the primitive heart tube. In the mouse, the organization, regulation and function of miR-1 and miR-133 have been analysed, i.e. cardiac transcription of miR-1/miR-133 bicistronic precursors is directly regulated by Mef2 and Srf.118 Additionally, experimental studies show that over-expression of miR-1 or miR-133 reduces the expression of Nkx-2.5 during differentiation of mouse embryonic stem cells.119 Expression of miR-126 is also detected in the pre-cardiac mesoderm in the developing chick,116 being observed later in the endocardium of the primitive heart tube and the vascular endothelium (Figure 6D). All these data suggest that micro-RNAs may be as important as transcription factors in controlling cardiac gene expression.
From the present review, a complete scenario of the genes involved in cardiac specification and in the early steps of heart formation can easily be analysed. The turning on and turning off of a particular gene or group of genes and the specification to different territories throughout development is clearly described and shown in Figures 3–5. This sequential and territorial relationship among genes is not presented according to gene family but by chronological order and with an indication of the final commitment of the cells expressing the gene. Although great progress has been made in order to produce cardiac cells from stem cells, knowledge of the genetic and therefore the molecular machinery involved in the formation of the heart is crucial to improve the production of myocardial cells safely and effectively.120 With this paper, we hope to fill the gap that is sometimes evident in the literature about the interplay of the genes that have a role in heart induction and in the early steps of cardiogenesis. This information could be used to design new therapeutic approaches based on a safe and precise differentiation of cardiogenic cells from any kind of stem cells.
Funding
This work was supported by the Spanish Ministry of Science and Innovation [BFU2007-66350/BFI to C.L.-S.]; and the Junta de Extremadura (FEDER) [CTS005 to V.G.-M.].
Acknowledgements
We are grateful to Dr I.S. Alvarez for comments and criticisms on the manuscript. We thank María Pérez for her invaluable technical help.
Conflict of interest: none declared.
References
Author notes
This article is part of the Spotlight Issue on: Cardiac Development